Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1405-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1405-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 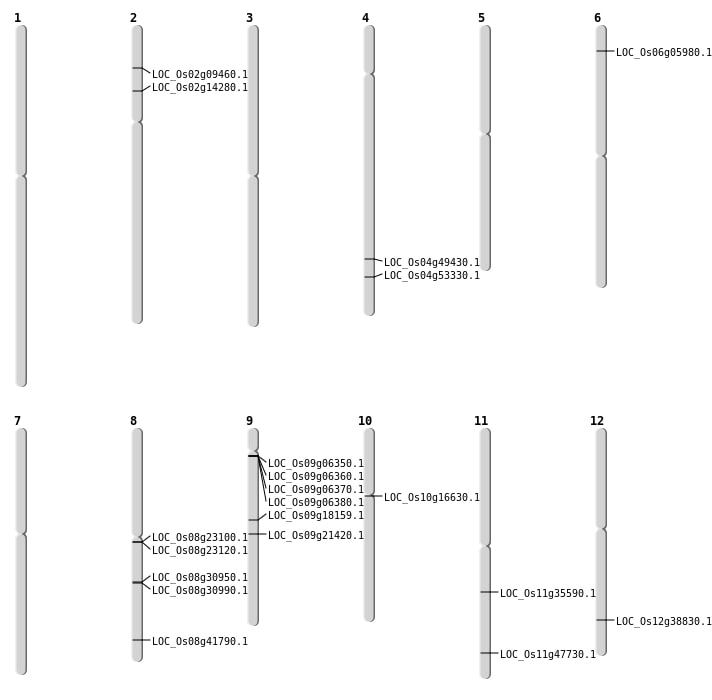
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11986787 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 17450265 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 19106605 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 21442377 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 22499904 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 4506032 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 8291484 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g16630 |
| SBS | Chr11 | 18226398 | G-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 20871147 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g35590 |
| SBS | Chr11 | 28685821 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 8714558 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 18920838 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 18920839 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 23860826 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g38830 |
| SBS | Chr12 | 2388292 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 5893467 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 7843195 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g14280 |
| SBS | Chr2 | 7843196 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g14280 |
| SBS | Chr3 | 5926907 | A-T | HOMO | INTRON | |
| SBS | Chr4 | 29497624 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g49430 |
| SBS | Chr5 | 26615447 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 24330371 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 27918525 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 3351195 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 9414937 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 26447007 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 5121394 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 12942598 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g21420 |
| SBS | Chr9 | 16321832 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 3094873 | C-T | HET | INTERGENIC |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 700797 | 700816 | 19 | |
| Deletion | Chr9 | 2928982 | 2928993 | 11 | |
| Deletion | Chr9 | 2978001 | 3008000 | 30000 | 4 |
| Deletion | Chr12 | 8446323 | 8446324 | 1 | |
| Deletion | Chr7 | 14380719 | 14380721 | 2 | |
| Deletion | Chr12 | 17037274 | 17037287 | 13 | |
| Deletion | Chr11 | 17087458 | 17087469 | 11 | |
| Deletion | Chr9 | 17365265 | 17365273 | 8 | |
| Deletion | Chr4 | 17657542 | 17657544 | 2 | |
| Deletion | Chr8 | 19136669 | 19136674 | 5 | LOC_Os08g30990 |
| Deletion | Chr9 | 20821382 | 20821389 | 7 | |
| Deletion | Chr1 | 25687860 | 25687865 | 5 | |
| Deletion | Chr8 | 26395118 | 26395138 | 20 | LOC_Os08g41790 |
| Deletion | Chr11 | 28689826 | 28690165 | 339 | |
| Deletion | Chr2 | 33003720 | 33003731 | 11 | |
| Deletion | Chr4 | 35093000 | 35093002 | 2 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 11895574 | 11895574 | 1 | |
| Insertion | Chr11 | 28794547 | 28794554 | 8 | LOC_Os11g47730 |
| Insertion | Chr4 | 19487614 | 19487614 | 1 | |
| Insertion | Chr6 | 11668089 | 11668090 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 163604 | 13675535 | |
| Inversion | Chr8 | 163655 | 13912724 | LOC_Os08g23100 |
| Inversion | Chr8 | 13930609 | 19107766 | 2 |
| Inversion | Chr8 | 13930789 | 19107766 | 2 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 163601 | Chr4 | 31765217 | LOC_Os04g53330 |
| Translocation | Chr9 | 11142067 | Chr4 | 31765549 | 2 |
| Translocation | Chr9 | 11144395 | Chr4 | 31765200 | LOC_Os04g53330 |
| Translocation | Chr8 | 13674878 | Chr2 | 4857905 | LOC_Os02g09460 |
| Translocation | Chr8 | 13675135 | Chr2 | 4857897 | LOC_Os02g09460 |
| Translocation | Chr8 | 13675528 | Chr6 | 2745486 | LOC_Os06g05980 |
| Translocation | Chr8 | 13912689 | Chr4 | 31765542 | 2 |
