Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1407-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1407-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 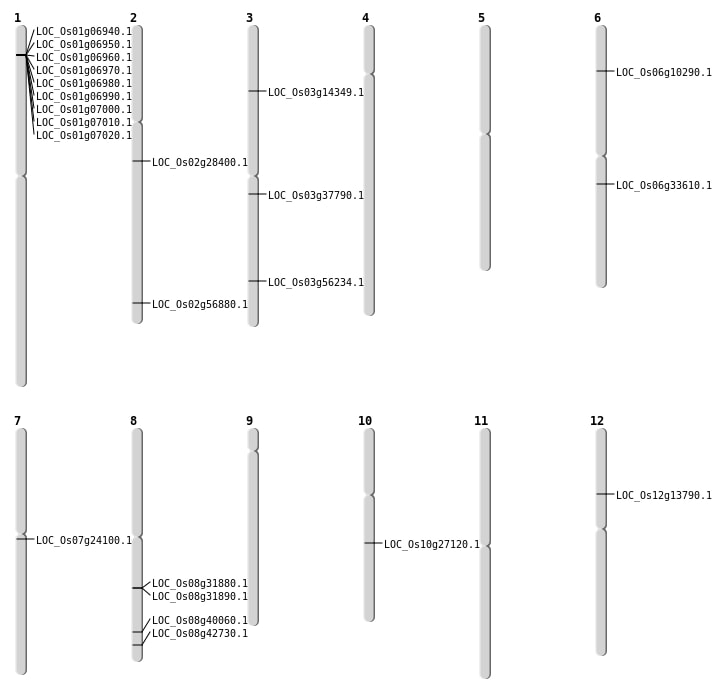
|
| Variant Information |
|---|
Single base substitutions: 26
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 32051386 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 12684313 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 7723928 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 16611014 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 26534058 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 16751100 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 35132292 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 6506846 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 6690970 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 32036328 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g56234 |
| SBS | Chr4 | 12796640 | G-T | Homo | INTRON | |
| SBS | Chr4 | 13161026 | T-C | Homo | INTERGENIC | |
| SBS | Chr4 | 15174338 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 16263205 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 19528332 | C-T | Homo | INTERGENIC | |
| SBS | Chr4 | 6106173 | T-C | Homo | INTERGENIC | |
| SBS | Chr4 | 7204239 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 14444785 | G-A | Homo | INTERGENIC | |
| SBS | Chr5 | 15664829 | G-C | Homo | INTERGENIC | |
| SBS | Chr5 | 15664843 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 22531661 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 5814876 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr7 | 13662741 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g24100 |
| SBS | Chr7 | 6938573 | T-G | HET | INTRON | |
| SBS | Chr8 | 17387945 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 17052502 | C-T | HET | INTERGENIC |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 1168967 | 1168969 | 2 | |
| Deletion | Chr1 | 1698957 | 1698959 | 2 | |
| Deletion | Chr7 | 1800797 | 1800810 | 13 | |
| Deletion | Chr1 | 2965486 | 2965492 | 6 | |
| Deletion | Chr1 | 3288527 | 3306523 | 17996 | 9 |
| Deletion | Chr11 | 3505052 | 3505105 | 53 | |
| Deletion | Chr6 | 5289013 | 5289019 | 6 | LOC_Os06g10290 |
| Deletion | Chr7 | 6963488 | 6963489 | 1 | |
| Deletion | Chr4 | 7270315 | 7270316 | 1 | |
| Deletion | Chr12 | 7798204 | 7798220 | 16 | LOC_Os12g13790 |
| Deletion | Chr3 | 7809010 | 7809017 | 7 | LOC_Os03g14349 |
| Deletion | Chr8 | 8163104 | 8163115 | 11 | |
| Deletion | Chr11 | 9588355 | 9588356 | 1 | |
| Deletion | Chr8 | 9949664 | 9949665 | 1 | |
| Deletion | Chr2 | 10240975 | 10240977 | 2 | |
| Deletion | Chr8 | 11581749 | 11581750 | 1 | |
| Deletion | Chr8 | 13907733 | 13907744 | 11 | |
| Deletion | Chr6 | 14137820 | 14137917 | 97 | |
| Deletion | Chr10 | 14293230 | 14293232 | 2 | LOC_Os10g27120 |
| Deletion | Chr8 | 14612265 | 14612271 | 6 | |
| Deletion | Chr12 | 16751094 | 16751095 | 1 | |
| Deletion | Chr2 | 16768178 | 16768292 | 114 | |
| Deletion | Chr2 | 16789098 | 16789102 | 4 | LOC_Os02g28400 |
| Deletion | Chr5 | 18371934 | 18371944 | 10 | |
| Deletion | Chr8 | 19763604 | 19766772 | 3168 | 2 |
| Deletion | Chr2 | 19962404 | 19962405 | 1 | |
| Deletion | Chr8 | 20307392 | 20307438 | 46 | |
| Deletion | Chr1 | 22203827 | 22203829 | 2 | |
| Deletion | Chr12 | 23487723 | 23487729 | 6 | |
| Deletion | Chr1 | 23959044 | 23959048 | 4 | |
| Deletion | Chr1 | 24533050 | 24533051 | 1 | |
| Deletion | Chr8 | 27034935 | 27034943 | 8 | LOC_Os08g42730 |
| Deletion | Chr3 | 31606958 | 31606960 | 2 | |
| Deletion | Chr2 | 34869724 | 34872238 | 2514 | LOC_Os02g56880 |
| Deletion | Chr3 | 35887579 | 35887585 | 6 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 17656179 | 17656193 | 15 | |
| Insertion | Chr10 | 15519735 | 15519788 | 54 | |
| Insertion | Chr3 | 25528254 | 25528274 | 21 | |
| Insertion | Chr5 | 4331899 | 4331899 | 1 | |
| Insertion | Chr5 | 5939695 | 5939744 | 50 | |
| Insertion | Chr7 | 4997471 | 4997474 | 4 | |
| Insertion | Chr8 | 25365864 | 25365864 | 1 | LOC_Os08g40060 |
| Insertion | Chr9 | 8986295 | 8986297 | 3 |
No Inversion
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 19565714 | Chr3 | 20953079 | LOC_Os06g33610 |
| Translocation | Chr6 | 19565726 | Chr3 | 20954558 | 2 |
