Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1410-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1410-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 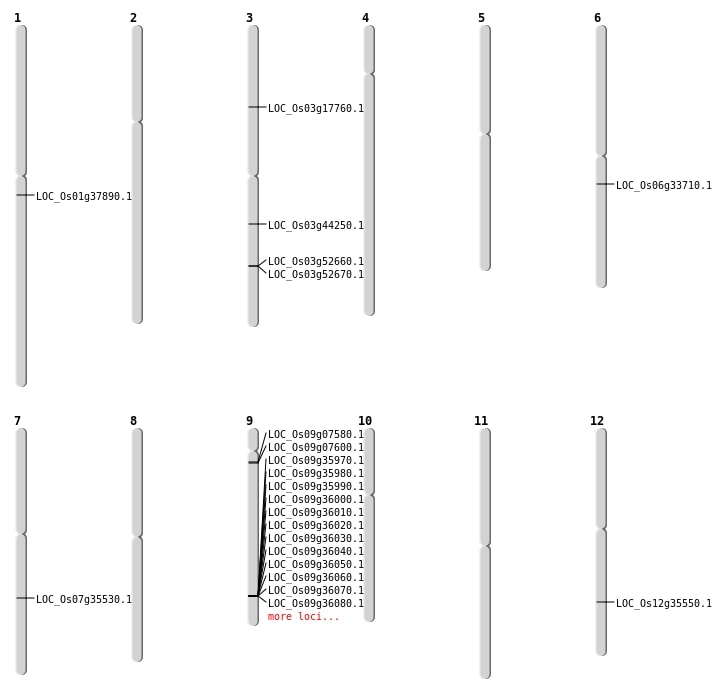
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19039256 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 21212464 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g37890 |
| SBS | Chr1 | 28578344 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 6924758 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 12724933 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 18189678 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 25587631 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 9988511 | G-A | HET | INTRON | |
| SBS | Chr12 | 23882110 | G-A | HET | INTRON | |
| SBS | Chr2 | 31223254 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 11397510 | C-T | HET | INTRON | |
| SBS | Chr3 | 23731131 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 4078694 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 7542899 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 22715379 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 27205600 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr4 | 6704485 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 12122345 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 2604852 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 12865294 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 17440556 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 17911557 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 5868777 | C-A | HET | INTRON | |
| SBS | Chr6 | 7756868 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 21263739 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g35530 |
| SBS | Chr7 | 26179850 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 27931129 | C-T | HET | INTRON | |
| SBS | Chr7 | 4161965 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 4671096 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 4866430 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 21714252 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 21840742 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 22762737 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 5808862 | T-C | HET | INTERGENIC |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 3065746 | 3065747 | 1 | |
| Deletion | Chr2 | 6092591 | 6092592 | 1 | |
| Deletion | Chr6 | 7243664 | 7243690 | 26 | |
| Deletion | Chr5 | 7883688 | 7883701 | 13 | |
| Deletion | Chr3 | 8335333 | 8335344 | 11 | |
| Deletion | Chr7 | 8388856 | 8388858 | 2 | |
| Deletion | Chr3 | 9880828 | 9880829 | 1 | LOC_Os03g17760 |
| Deletion | Chr1 | 10045254 | 10045260 | 6 | |
| Deletion | Chr12 | 10667449 | 10667459 | 10 | |
| Deletion | Chr1 | 11284425 | 11284426 | 1 | |
| Deletion | Chr2 | 15200382 | 15200384 | 2 | |
| Deletion | Chr10 | 16331542 | 16331543 | 1 | |
| Deletion | Chr9 | 20725001 | 20812000 | 87000 | 18 |
| Deletion | Chr12 | 21614856 | 21614877 | 21 | LOC_Os12g35550 |
| Deletion | Chr4 | 25505063 | 25505064 | 1 | |
| Deletion | Chr1 | 26907191 | 26907193 | 2 | |
| Deletion | Chr1 | 28177880 | 28177997 | 117 | |
| Deletion | Chr1 | 31053269 | 31053295 | 26 | |
| Deletion | Chr2 | 32835070 | 32835071 | 1 | |
| Deletion | Chr3 | 33937117 | 33937119 | 2 | |
| Deletion | Chr2 | 35649194 | 35649199 | 5 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 20915879 | 20915894 | 16 | |
| Insertion | Chr1 | 4011933 | 4011934 | 2 | |
| Insertion | Chr1 | 5671006 | 5671010 | 5 | |
| Insertion | Chr2 | 17953013 | 17953013 | 1 | |
| Insertion | Chr3 | 24866347 | 24866349 | 3 | LOC_Os03g44250 |
| Insertion | Chr3 | 25579459 | 25579460 | 2 | |
| Insertion | Chr3 | 35831458 | 35831458 | 1 | |
| Insertion | Chr4 | 29204549 | 29204549 | 1 | |
| Insertion | Chr8 | 13657406 | 13657407 | 2 | |
| Insertion | Chr8 | 5689988 | 5689988 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 4668678 | 4713124 | |
| Inversion | Chr3 | 11028908 | 11750193 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 3800664 | Chr6 | 19614551 | 2 |
| Translocation | Chr9 | 3811596 | Chr6 | 19614539 | 2 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr3 | 30195740 | 30198661 | 2921 | 2 |
