Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1420-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1420-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 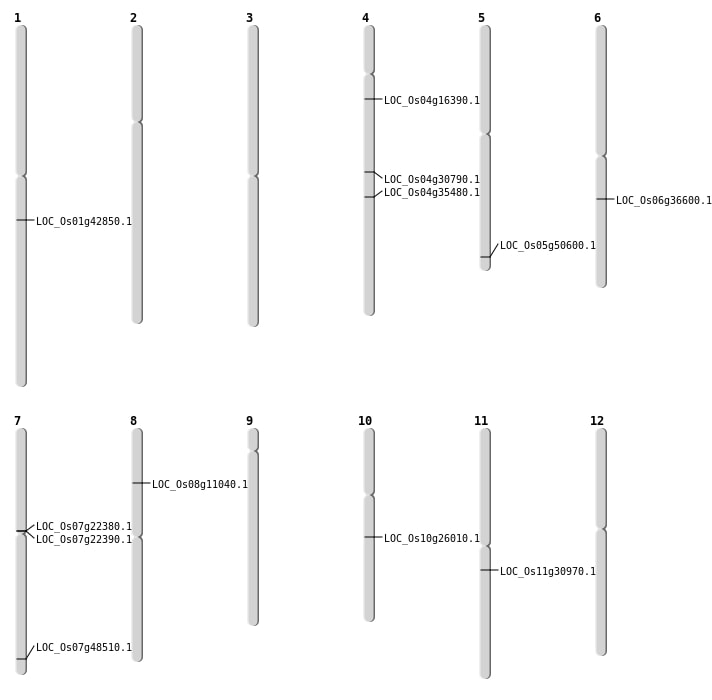
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17856347 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 32784143 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr1 | 32784144 | A-T | HET | UTR_5_PRIME | |
| SBS | Chr1 | 3811421 | C-T | HET | INTRON | |
| SBS | Chr1 | 41853803 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 14756579 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 19628958 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 16585888 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 18018852 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g30970 |
| SBS | Chr11 | 5852277 | T-A | HET | INTRON | |
| SBS | Chr11 | 9398032 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 9398033 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 5052223 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 16302295 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 20470517 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 14374711 | A-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 30277570 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 34745428 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 8907100 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g16390 |
| SBS | Chr5 | 17435107 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 22582515 | C-T | HET | INTRON | |
| SBS | Chr5 | 29004962 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g50600 |
| SBS | Chr6 | 11847071 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 20452563 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 23303022 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 27607436 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 28058903 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 8139991 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 11202939 | G-A | HET | INTRON | |
| SBS | Chr7 | 13043056 | C-A | HET | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1222530 | 1222531 | 1 | |
| Deletion | Chr4 | 2219633 | 2219634 | 1 | |
| Deletion | Chr3 | 4505898 | 4505899 | 1 | |
| Deletion | Chr2 | 5473664 | 5473668 | 4 | |
| Deletion | Chr11 | 5949750 | 5949756 | 6 | |
| Deletion | Chr3 | 8137690 | 8137692 | 2 | |
| Deletion | Chr1 | 9141175 | 9141176 | 1 | |
| Deletion | Chr4 | 10534917 | 10534918 | 1 | |
| Deletion | Chr7 | 12563369 | 12567516 | 4147 | 2 |
| Deletion | Chr10 | 13468157 | 13469781 | 1624 | LOC_Os10g26010 |
| Deletion | Chr3 | 14326903 | 14326904 | 1 | |
| Deletion | Chr2 | 15095468 | 15095469 | 1 | |
| Deletion | Chr5 | 15804515 | 15804516 | 1 | |
| Deletion | Chr4 | 18409024 | 18409026 | 2 | LOC_Os04g30790 |
| Deletion | Chr4 | 18453561 | 18453562 | 1 | |
| Deletion | Chr3 | 19108486 | 19108498 | 12 | |
| Deletion | Chr4 | 21593620 | 21593626 | 6 | LOC_Os04g35480 |
| Deletion | Chr6 | 23536984 | 23536988 | 4 | |
| Deletion | Chr1 | 24385371 | 24385376 | 5 | LOC_Os01g42850 |
| Deletion | Chr4 | 25498707 | 25498708 | 1 | |
| Deletion | Chr6 | 27748385 | 27748390 | 5 | |
| Deletion | Chr7 | 29012083 | 29012084 | 1 | LOC_Os07g48510 |
| Deletion | Chr2 | 32960773 | 32960774 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 2740150 | 2740150 | 1 | |
| Insertion | Chr2 | 29608488 | 29608492 | 5 | |
| Insertion | Chr6 | 21180561 | 21180561 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 6496873 | 7409535 | LOC_Os08g11040 |
| Inversion | Chr9 | 13168979 | 14004740 | |
| Inversion | Chr9 | 13168995 | 14004742 | |
| Inversion | Chr6 | 21507537 | 23340022 | LOC_Os06g36600 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 13468173 | Chr6 | 23337307 | LOC_Os10g26010 |
