Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1421-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1421-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 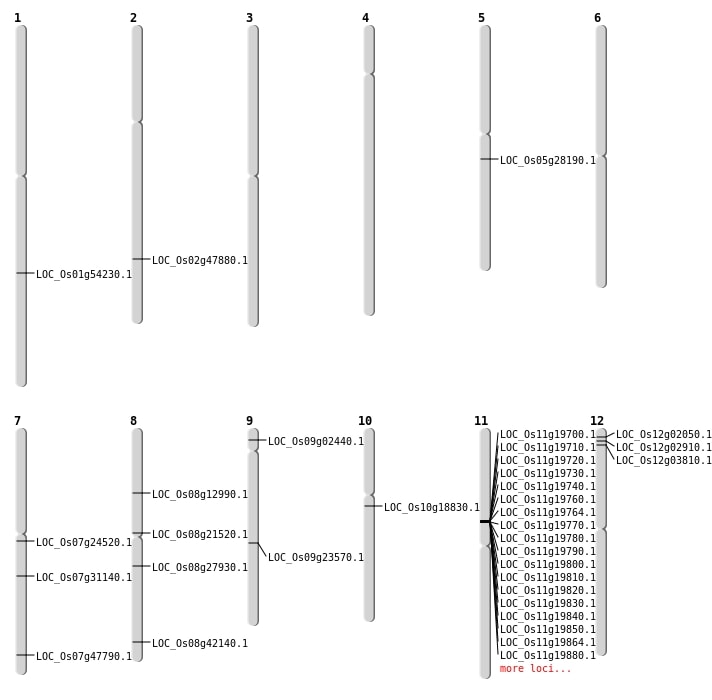
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1875765 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 20178634 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 28157171 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 31198289 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g54230 |
| SBS | Chr1 | 34905728 | A-T | HOMO | INTRON | |
| SBS | Chr10 | 753204 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 17078341 | C-T | HOMO | INTRON | |
| SBS | Chr11 | 22701136 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g38330 |
| SBS | Chr12 | 13211905 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 1763717 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 19548045 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 9460899 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 18944951 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 2185648 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 11560228 | C-A | HET | UTR_5_PRIME | |
| SBS | Chr4 | 1455503 | T-A | HET | INTRON | |
| SBS | Chr5 | 16503537 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g28190 |
| SBS | Chr5 | 21260574 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 28680674 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 5521975 | G-T | HET | INTRON | |
| SBS | Chr7 | 10046437 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 13144518 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 13957602 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g24520 |
| SBS | Chr7 | 15219379 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 28750574 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 5849558 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 12822436 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g21520 |
| SBS | Chr8 | 13004324 | C-A | HET | INTRON | |
| SBS | Chr8 | 17026009 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g27930 |
| SBS | Chr8 | 18228163 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 26630317 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g42140 |
| SBS | Chr8 | 26886163 | A-G | HOMO | INTRON | |
| SBS | Chr8 | 9121275 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 15905210 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 15968487 | C-G | HET | INTERGENIC |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 291266 | 291267 | 1 | |
| Deletion | Chr12 | 1555058 | 1555069 | 11 | LOC_Os12g03810 |
| Deletion | Chr8 | 7708765 | 7708770 | 5 | LOC_Os08g12990 |
| Deletion | Chr10 | 9585796 | 9585798 | 2 | LOC_Os10g18830 |
| Deletion | Chr11 | 11345001 | 11538000 | 193000 | 33 |
| Deletion | Chr10 | 12670173 | 12670175 | 2 | |
| Deletion | Chr5 | 13764361 | 13764363 | 2 | |
| Deletion | Chr11 | 15347001 | 15389000 | 42000 | 2 |
| Deletion | Chr7 | 18428958 | 18428963 | 5 | LOC_Os07g31140 |
| Deletion | Chr12 | 19524725 | 19524828 | 103 | |
| Deletion | Chr10 | 22041633 | 22041640 | 7 | |
| Deletion | Chr3 | 22272565 | 22272612 | 47 | |
| Deletion | Chr11 | 24750949 | 24750965 | 16 | LOC_Os11g41290 |
| Deletion | Chr3 | 25544230 | 25544235 | 5 | |
| Deletion | Chr2 | 26230100 | 26230118 | 18 | |
| Deletion | Chr8 | 26886063 | 26886089 | 26 | |
| Deletion | Chr5 | 28680675 | 28680676 | 1 | |
| Deletion | Chr2 | 29290843 | 29290856 | 13 | LOC_Os02g47880 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 1075382 | 1075383 | 2 | LOC_Os12g02910 |
| Insertion | Chr12 | 12451701 | 12451704 | 4 | |
| Insertion | Chr2 | 2937606 | 2937606 | 1 | |
| Insertion | Chr6 | 23951811 | 23951811 | 1 | |
| Insertion | Chr7 | 15852037 | 15852038 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 988104 | 1011468 | LOC_Os09g02440 |
| Inversion | Chr1 | 41538578 | 41688002 | |
| Inversion | Chr1 | 41688004 | 42096383 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 610773 | Chr4 | 4507458 | LOC_Os12g02050 |
| Translocation | Chr11 | 2352614 | Chr9 | 14008141 | LOC_Os09g23570 |
| Translocation | Chr11 | 21808432 | Chr7 | 28540481 | LOC_Os11g36960 |
| Translocation | Chr12 | 22158822 | Chr6 | 22062387 | |
| Translocation | Chr12 | 25698161 | Chr7 | 28540851 | LOC_Os07g47790 |
| Translocation | Chr12 | 25698163 | Chr7 | 28540853 | LOC_Os07g47790 |
