Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1422-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1422-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 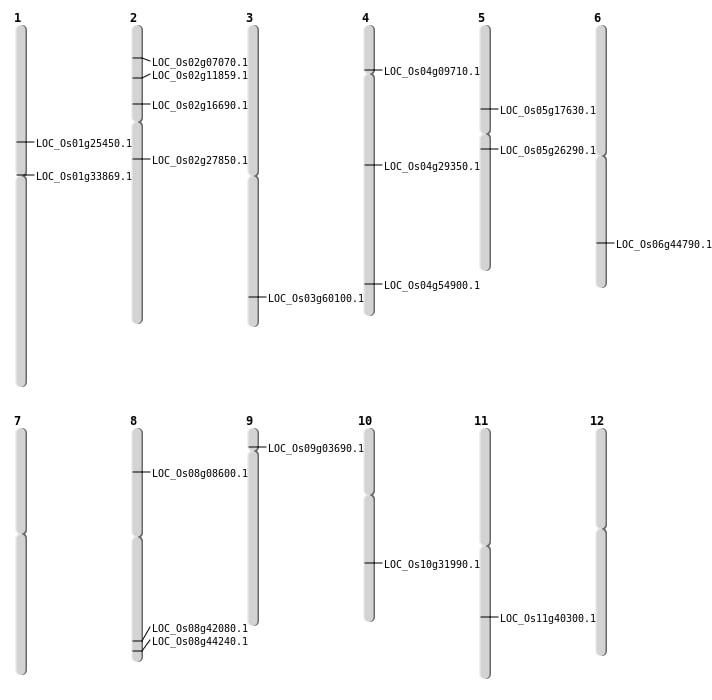
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14416269 | A-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g25450 |
| SBS | Chr1 | 14653220 | T-A | Homo | INTRON | |
| SBS | Chr1 | 18782848 | C-T | Homo | INTERGENIC | |
| SBS | Chr1 | 31360478 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr1 | 32143498 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 10306550 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 10742935 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 327580 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 5676749 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 6162982 | C-T | HET | INTRON | |
| SBS | Chr12 | 1445434 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 942259 | C-A | HET | INTRON | |
| SBS | Chr2 | 1380552 | C-A | HET | INTRON | |
| SBS | Chr2 | 15091241 | G-A | Homo | INTRON | |
| SBS | Chr2 | 19900701 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 35182622 | G-C | HET | INTRON | |
| SBS | Chr2 | 9540044 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g16690 |
| SBS | Chr3 | 16707129 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 10201046 | G-A | Homo | INTERGENIC | |
| SBS | Chr4 | 13911886 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 17434177 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g29350 |
| SBS | Chr4 | 32657789 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g54900 |
| SBS | Chr4 | 32657790 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g54900 |
| SBS | Chr4 | 5205191 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g09710 |
| SBS | Chr4 | 5205193 | G-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr5 | 27144432 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 28483827 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 3136765 | G-A | Homo | INTERGENIC | |
| SBS | Chr6 | 2576921 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 9151103 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 20691102 | C-T | Homo | UTR_3_PRIME | |
| SBS | Chr8 | 18273947 | G-A | Homo | INTERGENIC | |
| SBS | Chr8 | 27849641 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g44240 |
| SBS | Chr8 | 4976180 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g08600 |
| SBS | Chr9 | 3590549 | T-C | HET | INTRON |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 587265 | 587323 | 58 | |
| Deletion | Chr2 | 738728 | 738729 | 1 | |
| Deletion | Chr2 | 3630309 | 3630880 | 571 | LOC_Os02g07070 |
| Deletion | Chr4 | 5827841 | 5827849 | 8 | |
| Deletion | Chr11 | 7235867 | 7235891 | 24 | |
| Deletion | Chr5 | 10142531 | 10142547 | 16 | LOC_Os05g17630 |
| Deletion | Chr3 | 12559892 | 12559893 | 1 | |
| Deletion | Chr2 | 13017647 | 13017649 | 2 | |
| Deletion | Chr3 | 14406821 | 14406845 | 24 | |
| Deletion | Chr7 | 14994299 | 14994301 | 2 | |
| Deletion | Chr5 | 15291515 | 15291516 | 1 | LOC_Os05g26290 |
| Deletion | Chr9 | 15476626 | 15476646 | 20 | |
| Deletion | Chr8 | 16682241 | 16682256 | 15 | |
| Deletion | Chr5 | 17371174 | 17371176 | 2 | |
| Deletion | Chr8 | 20224195 | 20224196 | 1 | |
| Deletion | Chr6 | 20962290 | 20962291 | 1 | |
| Deletion | Chr9 | 22523811 | 22523816 | 5 | |
| Deletion | Chr11 | 24028847 | 24028861 | 14 | LOC_Os11g40300 |
| Deletion | Chr6 | 25090834 | 25090835 | 1 | |
| Deletion | Chr11 | 25292845 | 25292846 | 1 | |
| Deletion | Chr8 | 26572204 | 26572207 | 3 | LOC_Os08g42080 |
| Deletion | Chr6 | 27060331 | 27060345 | 14 | LOC_Os06g44790 |
| Deletion | Chr1 | 28173678 | 28173679 | 1 | |
| Deletion | Chr3 | 31582364 | 31582366 | 2 | |
| Deletion | Chr3 | 34176512 | 34176519 | 7 | LOC_Os03g60100 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 10763644 | 10763644 | 1 | |
| Insertion | Chr10 | 2478011 | 2478047 | 37 | |
| Insertion | Chr11 | 8388937 | 8388937 | 1 | |
| Insertion | Chr2 | 4931599 | 4931599 | 1 | |
| Insertion | Chr2 | 6146231 | 6146232 | 2 | LOC_Os02g11859 |
| Insertion | Chr7 | 27895935 | 27895961 | 27 | |
| Insertion | Chr7 | 28875945 | 28875945 | 1 | |
| Insertion | Chr9 | 9495466 | 9495467 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 22873883 | 23753384 | |
| Inversion | Chr4 | 22877268 | 23753386 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 1863460 | Chr2 | 16489487 | 2 |
| Translocation | Chr10 | 16807700 | Chr2 | 3630310 | 2 |
| Translocation | Chr10 | 16807709 | Chr1 | 18634626 | 2 |
