Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1427-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1427-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 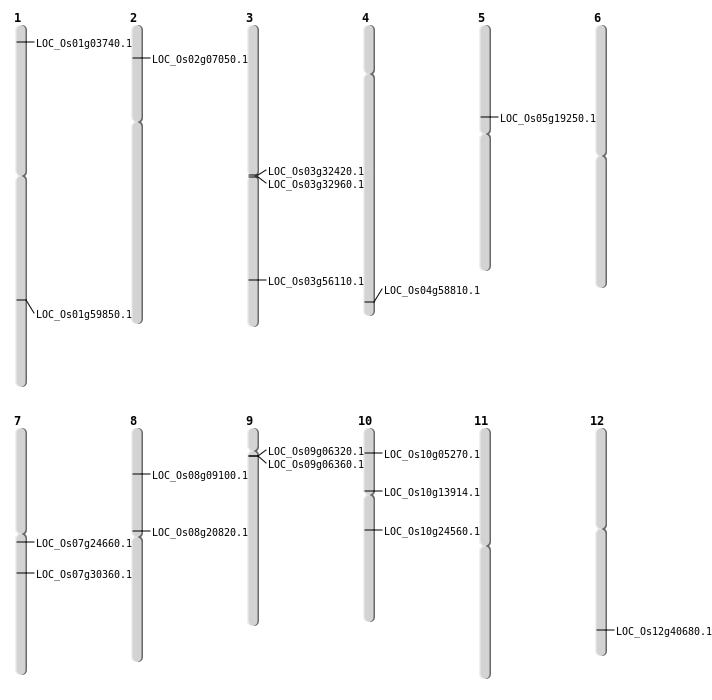
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1815428 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 37615119 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 12608016 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24560 |
| SBS | Chr10 | 6025997 | C-G | HOMO | INTRON | |
| SBS | Chr11 | 17692929 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 20306897 | T-C | HET | INTRON | |
| SBS | Chr11 | 21693739 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 26543839 | T-C | HET | INTRON | |
| SBS | Chr12 | 26095744 | T-C | HOMO | UTR_3_PRIME | |
| SBS | Chr2 | 12146797 | T-A | HOMO | INTRON | |
| SBS | Chr2 | 15000530 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 31095692 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 3616419 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g07050 |
| SBS | Chr3 | 18844208 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g32960 |
| SBS | Chr3 | 20269792 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 27608333 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 9206734 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 24625948 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 8171740 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 11184059 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g19250 |
| SBS | Chr6 | 10367951 | G-T | HET | INTRON | |
| SBS | Chr6 | 10367952 | A-G | HET | INTRON | |
| SBS | Chr6 | 10368176 | T-C | HET | INTRON | |
| SBS | Chr6 | 28526518 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 14029344 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g24660 |
| SBS | Chr7 | 17966317 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g30360 |
| SBS | Chr8 | 12495421 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g20820 |
| SBS | Chr8 | 17580535 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 2966624 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g06320 |
| SBS | Chr9 | 2992485 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g06360 |
| SBS | Chr9 | 3381797 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 4037892 | T-G | HOMO | SYNONYMOUS_CODING |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 1303950 | 1303963 | 13 | |
| Deletion | Chr1 | 1564742 | 1564744 | 2 | LOC_Os01g03740 |
| Deletion | Chr9 | 2968655 | 2968660 | 5 | |
| Deletion | Chr11 | 3153789 | 3153792 | 3 | |
| Deletion | Chr9 | 4447701 | 4447707 | 6 | |
| Deletion | Chr8 | 5267586 | 5271587 | 4001 | LOC_Os08g09100 |
| Deletion | Chr5 | 6634016 | 6634030 | 14 | |
| Deletion | Chr3 | 6926758 | 6926763 | 5 | |
| Deletion | Chr7 | 7685970 | 7686000 | 30 | |
| Deletion | Chr11 | 8963215 | 8963219 | 4 | |
| Deletion | Chr9 | 9823919 | 9823938 | 19 | |
| Deletion | Chr8 | 10553313 | 10553318 | 5 | |
| Deletion | Chr10 | 11051085 | 11051337 | 252 | |
| Deletion | Chr12 | 11602410 | 11602414 | 4 | |
| Deletion | Chr4 | 12990681 | 12990683 | 2 | |
| Deletion | Chr3 | 14576850 | 14576852 | 2 | |
| Deletion | Chr2 | 15772479 | 15772480 | 1 | |
| Deletion | Chr11 | 16138029 | 16138038 | 9 | |
| Deletion | Chr3 | 18539767 | 18539768 | 1 | LOC_Os03g32420 |
| Deletion | Chr8 | 19820347 | 19820354 | 7 | |
| Deletion | Chr2 | 20787325 | 20787335 | 10 | |
| Deletion | Chr3 | 21169076 | 21169086 | 10 | |
| Deletion | Chr3 | 21169082 | 21169092 | 10 | |
| Deletion | Chr6 | 21318003 | 21318009 | 6 | |
| Deletion | Chr1 | 24730909 | 24730911 | 2 | |
| Deletion | Chr8 | 25049345 | 25049348 | 3 | |
| Deletion | Chr12 | 25184783 | 25184786 | 3 | LOC_Os12g40680 |
| Deletion | Chr2 | 28095311 | 28095321 | 10 | |
| Deletion | Chr1 | 28108628 | 28108641 | 13 | |
| Deletion | Chr5 | 28352599 | 28352603 | 4 | |
| Deletion | Chr1 | 34618317 | 34618335 | 18 | LOC_Os01g59850 |
| Deletion | Chr4 | 34980745 | 34980764 | 19 | LOC_Os04g58810 |
| Deletion | Chr1 | 37000470 | 37000471 | 1 | |
| Deletion | Chr1 | 42803379 | 42803390 | 11 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 14032262 | 14032262 | 1 | |
| Insertion | Chr3 | 2470065 | 2470066 | 2 | |
| Insertion | Chr7 | 28750318 | 28750319 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 6963797 | 7558051 | LOC_Os10g13914 |
| Inversion | Chr11 | 14553782 | 21904033 | |
| Inversion | Chr3 | 31343894 | 31966086 | LOC_Os03g56110 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2607100 | Chr6 | 15332608 | LOC_Os10g05270 |
| Translocation | Chr9 | 6097346 | Chr8 | 5267590 |
