Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1430-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1430-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 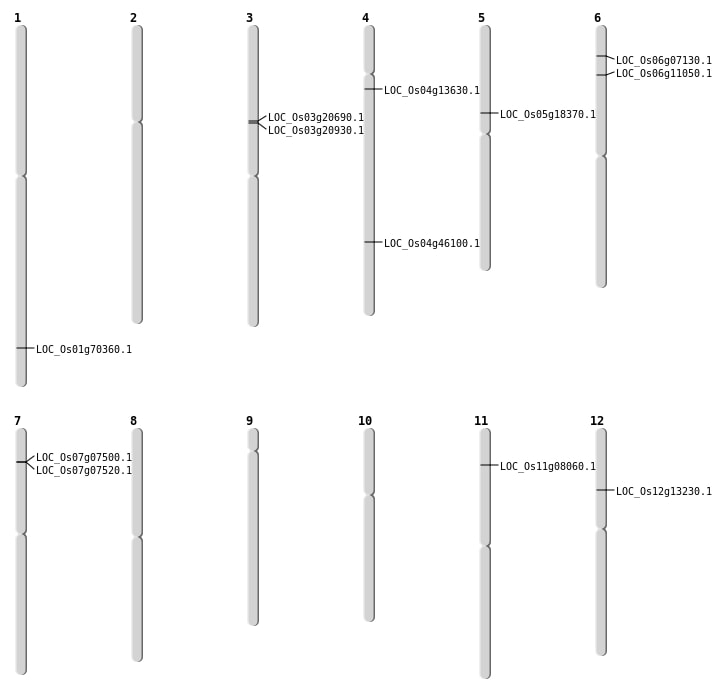
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12876381 | G-T | HET | INTRON | |
| SBS | Chr1 | 182401 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 38233378 | A-G | HET | INTRON | |
| SBS | Chr1 | 40763175 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g70360 |
| SBS | Chr10 | 21269311 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 2240093 | A-T | HET | INTRON | |
| SBS | Chr10 | 630628 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 16255082 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 17268542 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 26987705 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 28141436 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 7391556 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 10844427 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 11888574 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 13294485 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 14368391 | C-G | HET | INTRON | |
| SBS | Chr12 | 19447421 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 27268283 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 9381659 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 23962042 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 25539194 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 28461079 | C-T | HET | INTRON | |
| SBS | Chr3 | 22843833 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 25610973 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 33122697 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 35891990 | T-A | HOMO | INTRON | |
| SBS | Chr3 | 36114674 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 2252664 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 7606734 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g13630 |
| SBS | Chr5 | 10585579 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g18370 |
| SBS | Chr6 | 1987874 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 10608673 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 2307147 | A-T | HET | UTR_5_PRIME | |
| SBS | Chr7 | 2750554 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 1157425 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 1361376 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 22028049 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 3804896 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 6390334 | C-T | HOMO | INTRON |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 735815 | 735816 | 1 | |
| Deletion | Chr6 | 3416101 | 3416108 | 7 | LOC_Os06g07130 |
| Deletion | Chr7 | 3643476 | 3643477 | 1 | |
| Deletion | Chr8 | 3941103 | 3941104 | 1 | |
| Deletion | Chr11 | 4208263 | 4208267 | 4 | LOC_Os11g08060 |
| Deletion | Chr6 | 5791938 | 5791965 | 27 | LOC_Os06g11050 |
| Deletion | Chr12 | 7365042 | 7365043 | 1 | LOC_Os12g13230 |
| Deletion | Chr8 | 7384957 | 7384974 | 17 | |
| Deletion | Chr12 | 9583020 | 9583027 | 7 | |
| Deletion | Chr9 | 9777390 | 9777391 | 1 | |
| Deletion | Chr9 | 10517616 | 10517625 | 9 | |
| Deletion | Chr9 | 16408229 | 16408230 | 1 | |
| Deletion | Chr8 | 17255151 | 17255158 | 7 | |
| Deletion | Chr3 | 17279552 | 17279565 | 13 | |
| Deletion | Chr8 | 19419900 | 19419910 | 10 | |
| Deletion | Chr4 | 23530336 | 23530347 | 11 | |
| Deletion | Chr4 | 24129380 | 24129395 | 15 | |
| Deletion | Chr1 | 25527805 | 25527807 | 2 | |
| Deletion | Chr6 | 26635069 | 26635080 | 11 | |
| Deletion | Chr4 | 27322918 | 27322928 | 10 | LOC_Os04g46100 |
| Deletion | Chr5 | 28638904 | 28638909 | 5 | |
| Deletion | Chr5 | 29472664 | 29472666 | 2 | |
| Deletion | Chr3 | 36114653 | 36114666 | 13 | |
| Deletion | Chr1 | 38079538 | 38079549 | 11 | |
| Deletion | Chr1 | 42795279 | 42795307 | 28 |
Insertions: 6
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 3734283 | 3795654 | LOC_Os07g07500 |
| Inversion | Chr7 | 3748967 | 3795659 | LOC_Os07g07520 |
| Inversion | Chr3 | 11707145 | 11875810 | 2 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 9527620 | Chr6 | 25514514 | |
| Translocation | Chr10 | 23044590 | Chr7 | 3734287 | LOC_Os07g07500 |
| Translocation | Chr10 | 23044592 | Chr7 | 3749010 | LOC_Os07g07520 |
