Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1433-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1433-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 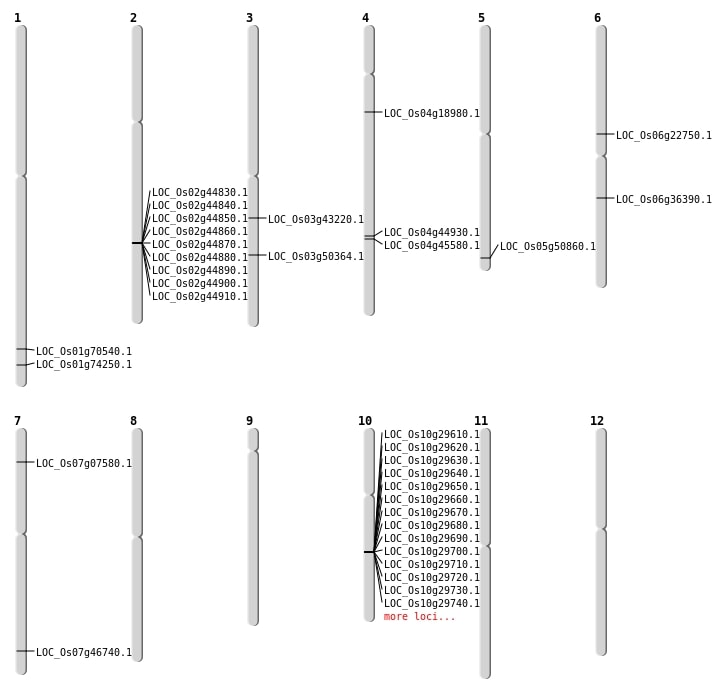
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 4903793 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 14044873 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 13877953 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 18746024 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 18856777 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 5573091 | A-G | HOMO | INTRON | |
| SBS | Chr12 | 18053197 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 19324841 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 11658179 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 18394737 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 10591478 | G-T | HOMO | UTR_3_PRIME | |
| SBS | Chr3 | 10719499 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 16260535 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 6506478 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 32838394 | G-A | HET | INTRON | |
| SBS | Chr5 | 14554857 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 17100762 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 25243184 | C-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 815971 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 815979 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 12239577 | T-A | HOMO | INTRON | |
| SBS | Chr6 | 12239578 | C-A | HOMO | INTRON | |
| SBS | Chr6 | 21893671 | A-G | HET | INTRON | |
| SBS | Chr6 | 23094866 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 11048634 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 11048637 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 11048638 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 18138939 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 15560136 | T-C | HET | INTERGENIC |
Deletions: 39
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 896613 | 896615 | 2 | |
| Deletion | Chr10 | 3382290 | 3382291 | 1 | |
| Deletion | Chr1 | 3473618 | 3473626 | 8 | |
| Deletion | Chr7 | 3798393 | 3798404 | 11 | LOC_Os07g07580 |
| Deletion | Chr12 | 4662517 | 4662518 | 1 | |
| Deletion | Chr2 | 6716242 | 6716253 | 11 | |
| Deletion | Chr5 | 7908379 | 7908380 | 1 | |
| Deletion | Chr8 | 9164003 | 9164011 | 8 | |
| Deletion | Chr7 | 9423356 | 9423359 | 3 | |
| Deletion | Chr6 | 10128471 | 10128473 | 2 | |
| Deletion | Chr7 | 11048081 | 11048083 | 2 | |
| Deletion | Chr8 | 11713806 | 11713808 | 2 | |
| Deletion | Chr6 | 13233037 | 13233046 | 9 | LOC_Os06g22750 |
| Deletion | Chr2 | 14048388 | 14048413 | 25 | |
| Deletion | Chr5 | 14945584 | 14945585 | 1 | |
| Deletion | Chr10 | 15391001 | 15953000 | 562000 | 93 |
| Deletion | Chr12 | 15850497 | 15850501 | 4 | |
| Deletion | Chr10 | 15966001 | 16428000 | 462000 | 53 |
| Deletion | Chr1 | 16141241 | 16141246 | 5 | |
| Deletion | Chr9 | 18231365 | 18231405 | 40 | |
| Deletion | Chr7 | 18706824 | 18706828 | 4 | |
| Deletion | Chr8 | 22208251 | 22208252 | 1 | |
| Deletion | Chr3 | 22324152 | 22324173 | 21 | |
| Deletion | Chr12 | 23337366 | 23337373 | 7 | |
| Deletion | Chr3 | 24102076 | 24102106 | 30 | LOC_Os03g43220 |
| Deletion | Chr5 | 24531774 | 24531779 | 5 | |
| Deletion | Chr12 | 25476006 | 25476051 | 45 | |
| Deletion | Chr4 | 26588382 | 26588383 | 1 | LOC_Os04g44930 |
| Deletion | Chr4 | 26962594 | 26962604 | 10 | LOC_Os04g45580 |
| Deletion | Chr2 | 27141001 | 27209000 | 68000 | 9 |
| Deletion | Chr4 | 27537352 | 27537354 | 2 | |
| Deletion | Chr1 | 27786411 | 27786422 | 11 | |
| Deletion | Chr7 | 27936990 | 27936993 | 3 | LOC_Os07g46740 |
| Deletion | Chr3 | 28744747 | 28744762 | 15 | LOC_Os03g50364 |
| Deletion | Chr5 | 29175523 | 29175533 | 10 | LOC_Os05g50860 |
| Deletion | Chr2 | 29856665 | 29856668 | 3 | |
| Deletion | Chr3 | 36223211 | 36223216 | 5 | |
| Deletion | Chr1 | 40857059 | 40857069 | 10 | LOC_Os01g70540 |
| Deletion | Chr1 | 43017202 | 43017207 | 5 | LOC_Os01g74250 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 28474874 | 28474875 | 2 | |
| Insertion | Chr4 | 10541808 | 10541828 | 21 | LOC_Os04g18980 |
| Insertion | Chr4 | 29415694 | 29415694 | 1 | |
| Insertion | Chr6 | 21372446 | 21372451 | 6 | LOC_Os06g36390 |
No Inversion
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 13699044 | Chr1 | 2503819 | |
| Translocation | Chr7 | 21189217 | Chr4 | 13727662 |
