Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1435-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1435-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 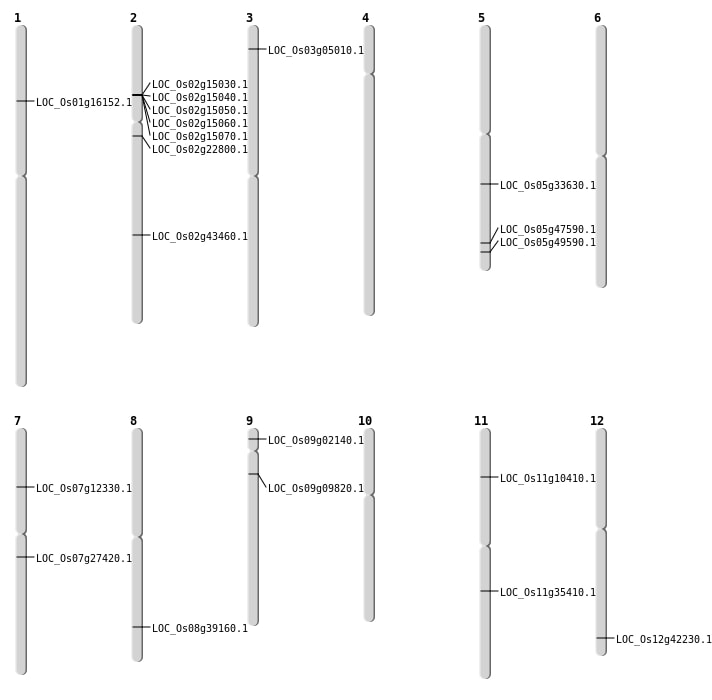
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 4646379 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 17896267 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 3730657 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 10258420 | C-T | HET | INTRON | |
| SBS | Chr12 | 12563046 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 26203000 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g42230 |
| SBS | Chr12 | 26638754 | A-G | HOMO | INTRON | |
| SBS | Chr2 | 10365641 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 13587366 | A-G | HET | INTRON | |
| SBS | Chr2 | 26855381 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 27396825 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 27791562 | C-T | HET | INTRON | |
| SBS | Chr3 | 2428373 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g05010 |
| SBS | Chr3 | 24432273 | T-C | HET | INTRON | |
| SBS | Chr4 | 27543579 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 19780691 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 19780692 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g33630 |
| SBS | Chr5 | 22491800 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr5 | 27264508 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g47590 |
| SBS | Chr6 | 14926255 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 2119994 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 7985 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 8791176 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 9448335 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 15976466 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g27420 |
| SBS | Chr7 | 26050079 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 2823943 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 795231 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 14667156 | T-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 5194750 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 11499875 | T-C | HET | INTRON | |
| SBS | Chr9 | 3866529 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 7638833 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 8026191 | G-A | HET | INTERGENIC |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 822581 | 822603 | 22 | LOC_Os09g02140 |
| Deletion | Chr5 | 1874944 | 1875026 | 82 | |
| Deletion | Chr3 | 2070942 | 2070945 | 3 | |
| Deletion | Chr3 | 2830184 | 2830186 | 2 | |
| Deletion | Chr2 | 3511115 | 3511144 | 29 | |
| Deletion | Chr1 | 3817334 | 3817342 | 8 | |
| Deletion | Chr3 | 4613683 | 4613695 | 12 | |
| Deletion | Chr9 | 5322521 | 5322522 | 1 | LOC_Os09g09820 |
| Deletion | Chr7 | 6959405 | 6959406 | 1 | LOC_Os07g12330 |
| Deletion | Chr2 | 8386450 | 8401663 | 15213 | 5 |
| Deletion | Chr11 | 8953756 | 8953820 | 64 | |
| Deletion | Chr1 | 9118162 | 9118167 | 5 | LOC_Os01g16152 |
| Deletion | Chr7 | 10578806 | 10578807 | 1 | |
| Deletion | Chr1 | 11098724 | 11098731 | 7 | |
| Deletion | Chr10 | 13664885 | 13664888 | 3 | |
| Deletion | Chr11 | 15263052 | 15263053 | 1 | |
| Deletion | Chr10 | 17819501 | 17819502 | 1 | |
| Deletion | Chr12 | 18262907 | 18262908 | 1 | |
| Deletion | Chr5 | 21933636 | 21933639 | 3 | |
| Deletion | Chr4 | 23834300 | 23834330 | 30 | |
| Deletion | Chr8 | 24734021 | 24734027 | 6 | LOC_Os08g39160 |
| Deletion | Chr1 | 25997410 | 25997413 | 3 | |
| Deletion | Chr2 | 26234643 | 26234665 | 22 | LOC_Os02g43460 |
| Deletion | Chr12 | 26638757 | 26638768 | 11 | |
| Deletion | Chr1 | 27674971 | 27674976 | 5 | |
| Deletion | Chr5 | 29192961 | 29192974 | 13 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 15086796 | 15086797 | 2 | |
| Insertion | Chr3 | 25891065 | 25891068 | 4 | |
| Insertion | Chr7 | 12108498 | 12108500 | 3 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 4458791 | 4461124 | |
| Inversion | Chr11 | 4729152 | 5651150 | LOC_Os11g10410 |
| Inversion | Chr11 | 4729165 | 5651169 | LOC_Os11g10410 |
| Inversion | Chr11 | 20751817 | 21578280 | LOC_Os11g35410 |
| Inversion | Chr2 | 27277087 | 27917565 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 10408739 | Chr4 | 17874145 | |
| Translocation | Chr11 | 16401942 | Chr1 | 6309094 | |
| Translocation | Chr11 | 16402202 | Chr1 | 6309004 | |
| Translocation | Chr12 | 21600405 | Chr6 | 14545839 | |
| Translocation | Chr12 | 24576978 | Chr2 | 13582794 | LOC_Os02g22800 |
| Translocation | Chr12 | 24577004 | Chr2 | 13587276 | LOC_Os02g22800 |
| Translocation | Chr12 | 24577005 | Chr2 | 13587199 | LOC_Os02g22800 |
| Translocation | Chr5 | 28448968 | Chr1 | 30432481 | LOC_Os05g49590 |
