Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1438-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1438-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 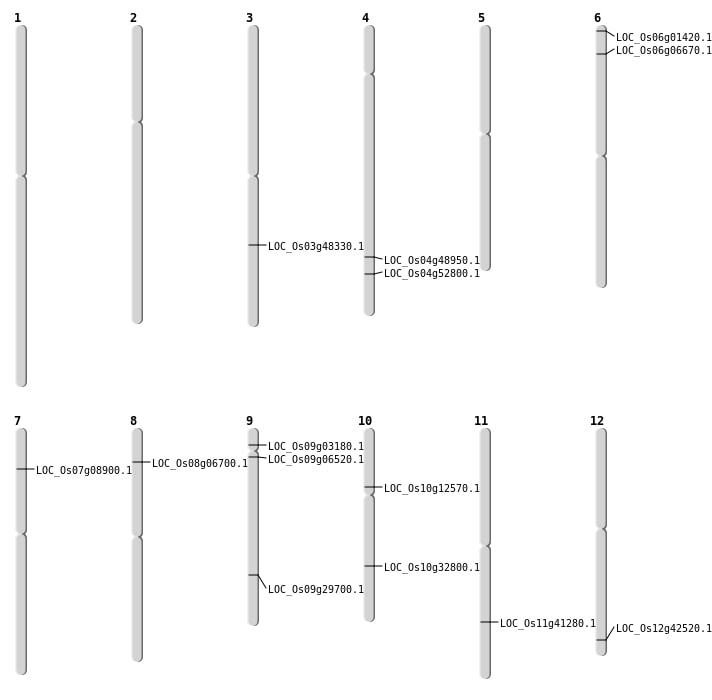
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10818765 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 27906078 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 39862620 | C-T | HET | INTRON | |
| SBS | Chr10 | 17176694 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g32800 |
| SBS | Chr10 | 20523759 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 6996656 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g12570 |
| SBS | Chr11 | 1354639 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 7786679 | C-T | HET | INTRON | |
| SBS | Chr3 | 1798736 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr3 | 27704366 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 16936020 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 2099783 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 28261886 | T-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 28261910 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 7226392 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 9359976 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 20591241 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 24497508 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 25676760 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 25987942 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 25987943 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 29486932 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 31210070 | T-A | HET | INTRON | |
| SBS | Chr7 | 14735549 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 15553849 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 2547471 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 2547472 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 3035356 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 4630759 | C-T | HET | STOP_GAINED | LOC_Os07g08900 |
| SBS | Chr8 | 23188055 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 3759235 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g06700 |
| SBS | Chr8 | 6570695 | T-G | HET | INTRON | |
| SBS | Chr9 | 17425455 | T-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 17718659 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 18060472 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g29700 |
| SBS | Chr9 | 3096957 | C-T | HET | STOP_GAINED | LOC_Os09g06520 |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 252032 | 252034 | 2 | LOC_Os06g01420 |
| Deletion | Chr9 | 1108975 | 1108976 | 1 | |
| Deletion | Chr9 | 1528161 | 1528162 | 1 | LOC_Os09g03180 |
| Deletion | Chr9 | 2083017 | 2083018 | 1 | |
| Deletion | Chr6 | 3132226 | 3132227 | 1 | LOC_Os06g06670 |
| Deletion | Chr8 | 3418549 | 3418688 | 139 | |
| Deletion | Chr6 | 3683418 | 3683419 | 1 | |
| Deletion | Chr5 | 4624642 | 4624643 | 1 | |
| Deletion | Chr6 | 5160073 | 5160074 | 1 | |
| Deletion | Chr6 | 6665112 | 6665286 | 174 | |
| Deletion | Chr6 | 7083214 | 7083231 | 17 | |
| Deletion | Chr4 | 7684398 | 7684400 | 2 | |
| Deletion | Chr3 | 11954178 | 11954179 | 1 | |
| Deletion | Chr5 | 19753249 | 19753279 | 30 | |
| Deletion | Chr3 | 21154135 | 21154144 | 9 | |
| Deletion | Chr10 | 21543517 | 21543521 | 4 | |
| Deletion | Chr12 | 21597035 | 21597038 | 3 | |
| Deletion | Chr12 | 26418032 | 26418037 | 5 | LOC_Os12g42520 |
| Deletion | Chr3 | 26818253 | 26818259 | 6 | |
| Deletion | Chr4 | 29196311 | 29196312 | 1 | LOC_Os04g48950 |
| Deletion | Chr4 | 31435706 | 31435721 | 15 | LOC_Os04g52800 |
| Deletion | Chr4 | 31690794 | 31690795 | 1 | |
| Deletion | Chr3 | 32126714 | 32126728 | 14 | |
| Deletion | Chr2 | 33419553 | 33419567 | 14 | |
| Deletion | Chr3 | 35718082 | 35718086 | 4 |
No Insertion
No Inversion
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 24746994 | Chr3 | 27526894 | 2 |
