Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1440-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1440-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 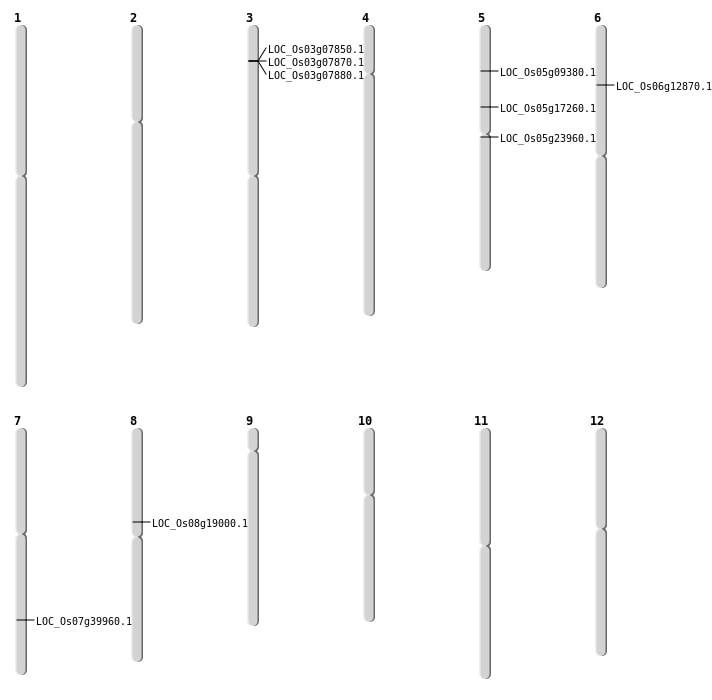
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22492453 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 31799305 | T-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 36296449 | A-T | HOMO | INTRON | |
| SBS | Chr10 | 1696295 | A-T | HET | INTRON | |
| SBS | Chr10 | 8431463 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 10377103 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 23211714 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 116227 | T-A | HET | INTRON | |
| SBS | Chr12 | 16706539 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr12 | 18349727 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 21244169 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 13265423 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 32970450 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 12705674 | C-T | HET | INTRON | |
| SBS | Chr3 | 23602268 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 5696964 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 20301222 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 13795372 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g23960 |
| SBS | Chr5 | 23612939 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 27013296 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 4496151 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 9877655 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g17260 |
| SBS | Chr6 | 7044408 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g12870 |
| SBS | Chr7 | 23976478 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g39960 |
| SBS | Chr8 | 11340104 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g19000 |
| SBS | Chr8 | 1351532 | A-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 16030401 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 27671527 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr8 | 316294 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 16005060 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 4167554 | C-T | HET | INTRON | |
| SBS | Chr9 | 6589662 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 759600 | T-A | HET | INTRON |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 540709 | 540712 | 3 | |
| Deletion | Chr5 | 923318 | 923322 | 4 | |
| Deletion | Chr11 | 1221698 | 1221705 | 7 | |
| Deletion | Chr10 | 1626212 | 1626228 | 16 | |
| Deletion | Chr4 | 2587260 | 2587273 | 13 | |
| Deletion | Chr6 | 2994625 | 2994636 | 11 | |
| Deletion | Chr3 | 3994517 | 4009447 | 14930 | 3 |
| Deletion | Chr3 | 4549450 | 4550175 | 725 | |
| Deletion | Chr1 | 5108374 | 5108382 | 8 | |
| Deletion | Chr5 | 5269894 | 5269903 | 9 | LOC_Os05g09380 |
| Deletion | Chr5 | 10935699 | 10935715 | 16 | |
| Deletion | Chr10 | 10965444 | 10965468 | 24 | |
| Deletion | Chr5 | 12253823 | 12253829 | 6 | |
| Deletion | Chr12 | 12880784 | 12880785 | 1 | |
| Deletion | Chr7 | 21433022 | 21433027 | 5 | |
| Deletion | Chr9 | 22518536 | 22518537 | 1 | |
| Deletion | Chr9 | 22518540 | 22518541 | 1 | |
| Deletion | Chr4 | 22556692 | 22556699 | 7 | |
| Deletion | Chr3 | 25226661 | 25226664 | 3 | |
| Deletion | Chr4 | 26354613 | 26354620 | 7 | |
| Deletion | Chr5 | 27987000 | 27987006 | 6 | |
| Deletion | Chr4 | 29329271 | 29329278 | 7 | |
| Deletion | Chr2 | 33468691 | 33468692 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 36921330 | 36921331 | 2 | |
| Insertion | Chr12 | 3070387 | 3070388 | 2 | |
| Insertion | Chr12 | 6749362 | 6749362 | 1 | |
| Insertion | Chr7 | 11351622 | 11351624 | 3 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 13689707 | 13754533 |
No Translocation
