Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1442-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1442-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 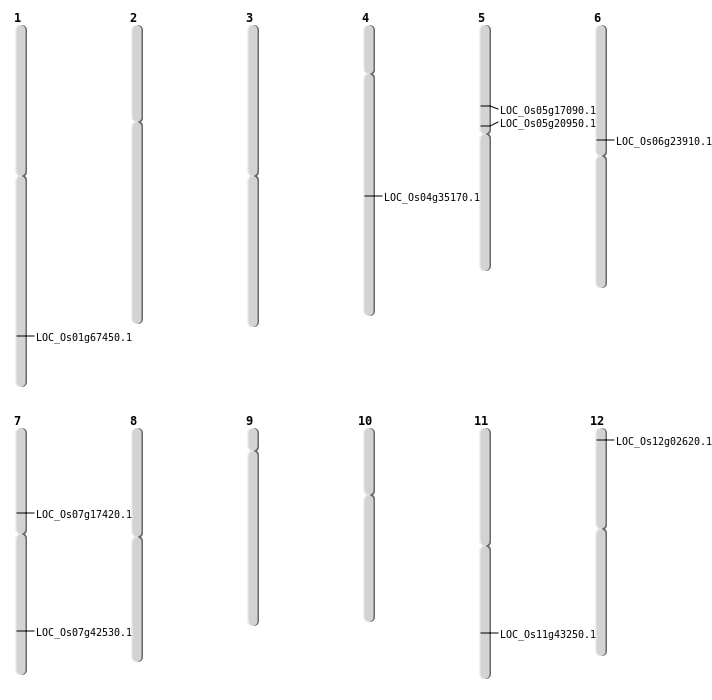
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15807549 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 19829022 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 36804635 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 38572294 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 8689271 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 13054570 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 21552861 | A-T | HOMO | INTRON | |
| SBS | Chr11 | 26089589 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g43250 |
| SBS | Chr11 | 5544898 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 13894111 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 26194760 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr12 | 26568237 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 8234326 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 908036 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g02620 |
| SBS | Chr12 | 9957165 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 15479254 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 21370549 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 11986910 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 13502297 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 22433082 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 26744661 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 14119674 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 21374216 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g35170 |
| SBS | Chr4 | 2738117 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 2738118 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 455276 | A-T | HET | INTRON | |
| SBS | Chr4 | 4921975 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 9741525 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 12320560 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g20950 |
| SBS | Chr5 | 18923706 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 13971698 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g23910 |
| SBS | Chr7 | 17505771 | T-C | HOMO | INTRON | |
| SBS | Chr7 | 19123782 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 4345132 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 9914621 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 12248210 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 1816990 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 3989692 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 8020293 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 21567511 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 21567531 | C-T | HET | INTERGENIC |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 3060272 | 3060287 | 15 | |
| Deletion | Chr8 | 4327327 | 4327330 | 3 | |
| Deletion | Chr11 | 5411477 | 5411479 | 2 | |
| Deletion | Chr8 | 9382316 | 9382321 | 5 | |
| Deletion | Chr5 | 9756581 | 9756587 | 6 | LOC_Os05g17090 |
| Deletion | Chr6 | 10794197 | 10794213 | 16 | |
| Deletion | Chr6 | 14323409 | 14323420 | 11 | |
| Deletion | Chr9 | 16649614 | 16649627 | 13 | |
| Deletion | Chr9 | 17351664 | 17351675 | 11 | |
| Deletion | Chr1 | 17886773 | 17887008 | 235 | |
| Deletion | Chr5 | 20022461 | 20022473 | 12 | |
| Deletion | Chr4 | 21408230 | 21408231 | 1 | |
| Deletion | Chr5 | 24046507 | 24046508 | 1 | |
| Deletion | Chr7 | 25452971 | 25452988 | 17 | LOC_Os07g42530 |
| Deletion | Chr6 | 26149694 | 26149697 | 3 | |
| Deletion | Chr3 | 31146758 | 31146768 | 10 | |
| Deletion | Chr4 | 35363892 | 35363893 | 1 | |
| Deletion | Chr1 | 39194575 | 39201652 | 7077 | LOC_Os01g67450 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 28012238 | 28012239 | 2 | |
| Insertion | Chr5 | 28034317 | 28034317 | 1 | |
| Insertion | Chr7 | 10285652 | 10285653 | 2 | LOC_Os07g17420 |
| Insertion | Chr7 | 24851127 | 24851130 | 4 | |
| Insertion | Chr8 | 16396233 | 16396258 | 26 | |
| Insertion | Chr8 | 9564202 | 9564202 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 15691180 | 15877600 | |
| Inversion | Chr9 | 17111631 | 17174114 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 8941254 | Chr1 | 30237189 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr3 | 26151514 | 26151606 | 92 |
