Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1448-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1448-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 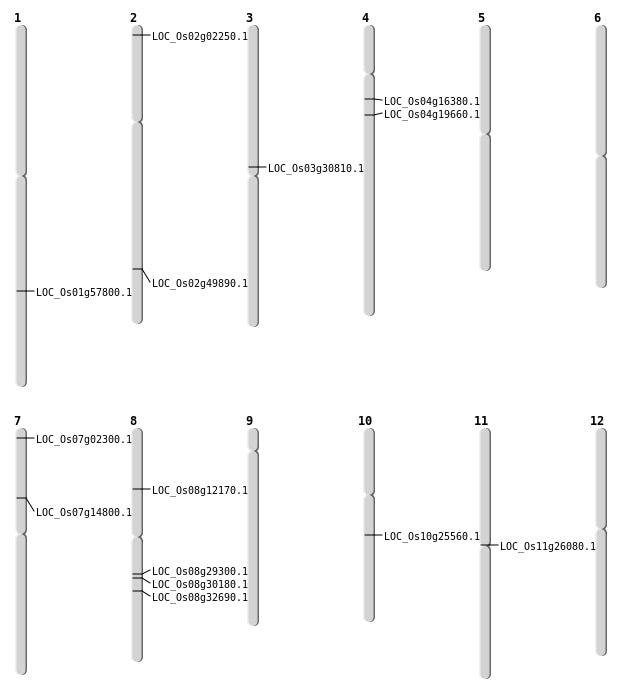
|
| Variant Information |
|---|
Single base substitutions: 51
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 25668902 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 25692914 | C-A | HET | INTRON | |
| SBS | Chr1 | 26901884 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 31781842 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 32446477 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 10931559 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 11852717 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 13721016 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 1589648 | G-A | HET | INTRON | |
| SBS | Chr10 | 17461233 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 14918880 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g26080 |
| SBS | Chr11 | 19196870 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 8060660 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 8342691 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 9909122 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 16938955 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 23515330 | T-C | HET | INTRON | |
| SBS | Chr12 | 25421512 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 18415471 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 30484755 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g49890 |
| SBS | Chr3 | 17447518 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 17565820 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g30810 |
| SBS | Chr3 | 18710991 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 23666282 | C-G | HET | INTRON | |
| SBS | Chr3 | 24563378 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 10970459 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g19660 |
| SBS | Chr4 | 20320235 | G-T | HOMO | INTRON | |
| SBS | Chr4 | 21067445 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 34594860 | T-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 8903060 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g16380 |
| SBS | Chr5 | 10413133 | G-A | HET | INTRON | |
| SBS | Chr6 | 21274818 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 26391046 | C-T | HET | INTRON | |
| SBS | Chr6 | 26524001 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 29885685 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 19394143 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 22798952 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr7 | 23436823 | C-T | HET | INTRON | |
| SBS | Chr7 | 28678909 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 3523889 | G-A | HET | INTRON | |
| SBS | Chr7 | 8448530 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g14800 |
| SBS | Chr8 | 17977958 | G-A | HET | STOP_GAINED | LOC_Os08g29300 |
| SBS | Chr8 | 18554245 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g30180 |
| SBS | Chr8 | 22383907 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 2557804 | T-A | HOMO | INTRON | |
| SBS | Chr8 | 7168202 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g12170 |
| SBS | Chr8 | 8558799 | A-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 16217871 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 4091276 | C-T | HOMO | INTRON | |
| SBS | Chr9 | 5471080 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 5832400 | C-T | HOMO | INTERGENIC |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 78894 | 78907 | 13 | |
| Deletion | Chr2 | 728886 | 728892 | 6 | LOC_Os02g02250 |
| Deletion | Chr5 | 4184431 | 4184441 | 10 | |
| Deletion | Chr12 | 4464131 | 4464134 | 3 | |
| Deletion | Chr3 | 6762215 | 6762217 | 2 | |
| Deletion | Chr1 | 7404681 | 7404689 | 8 | |
| Deletion | Chr10 | 8341257 | 8341269 | 12 | |
| Deletion | Chr11 | 10381157 | 10381159 | 2 | |
| Deletion | Chr10 | 13231873 | 13231875 | 2 | LOC_Os10g25560 |
| Deletion | Chr9 | 13605194 | 13605212 | 18 | |
| Deletion | Chr10 | 14917489 | 14917520 | 31 | |
| Deletion | Chr12 | 23026298 | 23026304 | 6 | |
| Deletion | Chr8 | 23818926 | 23818932 | 6 | |
| Deletion | Chr6 | 28845728 | 28845760 | 32 | |
| Deletion | Chr2 | 30527995 | 30527996 | 1 | |
| Deletion | Chr6 | 31029423 | 31029425 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr4 | 4266037 | 4266038 | 2 | |
| Insertion | Chr7 | 761162 | 761162 | 1 | LOC_Os07g02300 |
| Insertion | Chr8 | 16438848 | 16438855 | 8 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 33432888 | 33684589 | LOC_Os01g57800 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 24604183 | Chr8 | 20252392 | LOC_Os08g32690 |
