Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1450-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1450-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 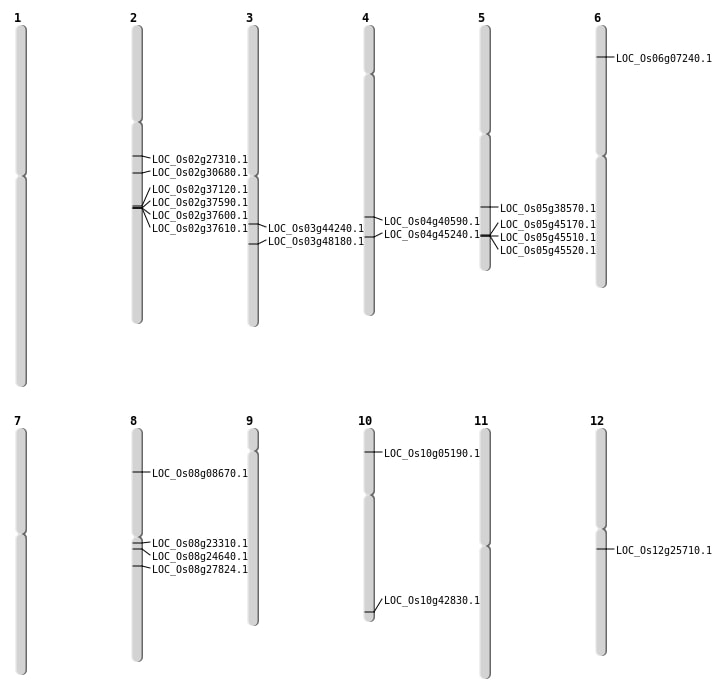
|
| Variant Information |
|---|
Single base substitutions: 26
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 31563020 | T-G | HET | INTRON | |
| SBS | Chr10 | 1652204 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 18306693 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 23095729 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g42830 |
| SBS | Chr10 | 3176816 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 14046243 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 20050670 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 13767444 | A-T | HET | INTRON | |
| SBS | Chr12 | 19488898 | T-A | HET | INTRON | |
| SBS | Chr2 | 12673511 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 16128118 | A-C | Homo | INTERGENIC | |
| SBS | Chr2 | 20337925 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 21299431 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 21338992 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 4780233 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 13696780 | C-T | Homo | INTERGENIC | |
| SBS | Chr4 | 20299760 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 26791299 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 26224237 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g45170 |
| SBS | Chr5 | 8386245 | G-T | Homo | INTERGENIC | |
| SBS | Chr5 | 8640067 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 14070059 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g23310 |
| SBS | Chr8 | 23642063 | A-C | Homo | INTERGENIC | |
| SBS | Chr8 | 4289995 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 4289996 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 5194750 | G-A | HET | INTERGENIC |
Deletions: 52
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 182607 | 182612 | 5 | |
| Deletion | Chr1 | 723520 | 723521 | 1 | |
| Deletion | Chr11 | 1339527 | 1339534 | 7 | |
| Deletion | Chr8 | 2618311 | 2618312 | 1 | |
| Deletion | Chr6 | 3469880 | 3469890 | 10 | LOC_Os06g07240 |
| Deletion | Chr3 | 4054484 | 4054491 | 7 | |
| Deletion | Chr12 | 4756709 | 4756710 | 1 | |
| Deletion | Chr6 | 4825514 | 4825515 | 1 | |
| Deletion | Chr8 | 5020108 | 5020116 | 8 | LOC_Os08g08670 |
| Deletion | Chr4 | 5253034 | 5253035 | 1 | |
| Deletion | Chr1 | 5483257 | 5483258 | 1 | |
| Deletion | Chr12 | 7688311 | 7688315 | 4 | |
| Deletion | Chr3 | 9303454 | 9303458 | 4 | |
| Deletion | Chr10 | 9779510 | 9779517 | 7 | |
| Deletion | Chr2 | 10185620 | 10185634 | 14 | |
| Deletion | Chr1 | 10596222 | 10596223 | 1 | |
| Deletion | Chr10 | 11887008 | 11887009 | 1 | |
| Deletion | Chr3 | 12226563 | 12226564 | 1 | |
| Deletion | Chr8 | 12814398 | 12814399 | 1 | |
| Deletion | Chr1 | 13618032 | 13618052 | 20 | |
| Deletion | Chr11 | 14744328 | 14744334 | 6 | |
| Deletion | Chr11 | 14866277 | 14866278 | 1 | |
| Deletion | Chr12 | 14897338 | 14897361 | 23 | LOC_Os12g25710 |
| Deletion | Chr7 | 14909655 | 14909659 | 4 | |
| Deletion | Chr2 | 16093881 | 16093884 | 3 | LOC_Os02g27310 |
| Deletion | Chr11 | 16711936 | 16711939 | 3 | |
| Deletion | Chr8 | 16951770 | 16951778 | 8 | LOC_Os08g27824 |
| Deletion | Chr2 | 17701939 | 17701940 | 1 | |
| Deletion | Chr2 | 18278740 | 18278744 | 4 | LOC_Os02g30680 |
| Deletion | Chr11 | 18674862 | 18674866 | 4 | |
| Deletion | Chr10 | 19464993 | 19464998 | 5 | |
| Deletion | Chr7 | 21106305 | 21106311 | 6 | |
| Deletion | Chr10 | 21512998 | 21513025 | 27 | |
| Deletion | Chr8 | 21744799 | 21744803 | 4 | |
| Deletion | Chr8 | 21834767 | 21834773 | 6 | |
| Deletion | Chr5 | 22626968 | 22627001 | 33 | LOC_Os05g38570 |
| Deletion | Chr2 | 22691325 | 22697918 | 6593 | 3 |
| Deletion | Chr1 | 22753305 | 22753317 | 12 | |
| Deletion | Chr2 | 22782000 | 22782007 | 7 | |
| Deletion | Chr4 | 24102042 | 24102044 | 2 | LOC_Os04g40590 |
| Deletion | Chr3 | 24859346 | 24859360 | 14 | LOC_Os03g44240 |
| Deletion | Chr8 | 25769465 | 25769471 | 6 | |
| Deletion | Chr1 | 25783936 | 25783940 | 4 | |
| Deletion | Chr5 | 26386454 | 26390446 | 3992 | 2 |
| Deletion | Chr4 | 26727371 | 26727380 | 9 | LOC_Os04g45240 |
| Deletion | Chr3 | 27272952 | 27272963 | 11 | |
| Deletion | Chr5 | 27293129 | 27293131 | 2 | |
| Deletion | Chr2 | 27367449 | 27367451 | 2 | |
| Deletion | Chr3 | 27413538 | 27413562 | 24 | LOC_Os03g48180 |
| Deletion | Chr6 | 29028536 | 29030975 | 2439 | |
| Deletion | Chr1 | 30982912 | 30982913 | 1 | |
| Deletion | Chr1 | 31140317 | 31140325 | 8 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 19701411 | 19701411 | 1 | |
| Insertion | Chr10 | 2543902 | 2543934 | 33 | LOC_Os10g05190 |
| Insertion | Chr3 | 22561110 | 22561111 | 2 | |
| Insertion | Chr3 | 24650290 | 24650292 | 3 | |
| Insertion | Chr5 | 5462300 | 5462301 | 2 | |
| Insertion | Chr6 | 18627978 | 18627978 | 1 | |
| Insertion | Chr6 | 24945477 | 24945486 | 10 | |
| Insertion | Chr8 | 14893546 | 14893546 | 1 | LOC_Os08g24640 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 22440753 | 22926030 | LOC_Os02g37120 |
| Inversion | Chr2 | 22440795 | 22691326 | 2 |
| Inversion | Chr5 | 24158844 | 24345999 | |
| Inversion | Chr5 | 24158881 | 24346036 |
No Translocation
