Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1452-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1452-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 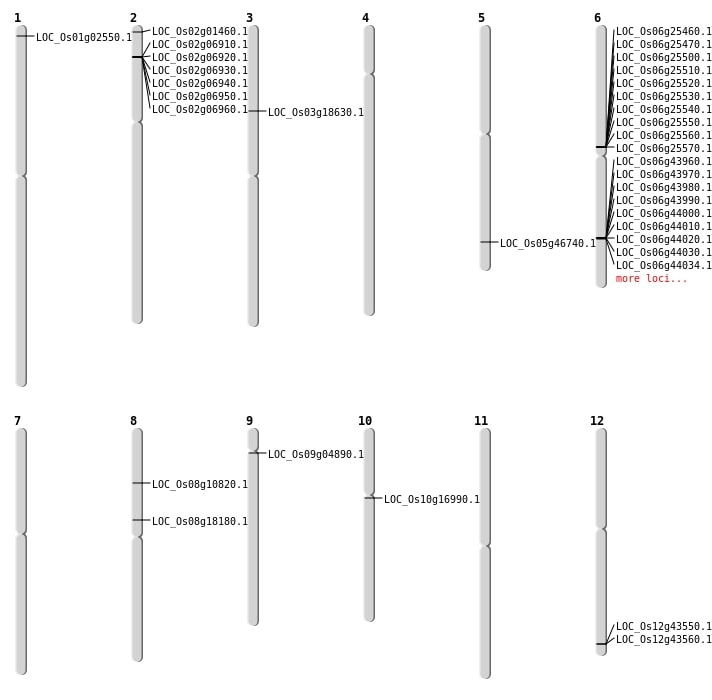
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11958830 | G-T | HET | INTRON | |
| SBS | Chr1 | 28151786 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 18758577 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 6523880 | G-A | HET | INTRON | |
| SBS | Chr12 | 13173457 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 9711273 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 13884176 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 4134343 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 4219768 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 9563474 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 10438218 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g18630 |
| SBS | Chr3 | 11097630 | G-C | HET | INTRON | |
| SBS | Chr3 | 11329453 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 18632069 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 10426458 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 18313549 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 12084475 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 12084477 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 29131839 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 4745359 | T-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 9038607 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 14906686 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 22234755 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 3594125 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 6371845 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g10820 |
| SBS | Chr8 | 9799919 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 20677536 | G-A | HET | INTRON | |
| SBS | Chr9 | 2615470 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g04890 |
| SBS | Chr9 | 749557 | T-A | HET | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 271065 | 271072 | 7 | LOC_Os02g01460 |
| Deletion | Chr11 | 734069 | 734070 | 1 | |
| Deletion | Chr1 | 841775 | 841787 | 12 | LOC_Os01g02550 |
| Deletion | Chr7 | 1588066 | 1588067 | 1 | |
| Deletion | Chr2 | 3475867 | 3475876 | 9 | |
| Deletion | Chr4 | 3476308 | 3476313 | 5 | |
| Deletion | Chr2 | 3479001 | 3541000 | 62000 | 6 |
| Deletion | Chr7 | 4419431 | 4419439 | 8 | |
| Deletion | Chr8 | 5867030 | 5867036 | 6 | |
| Deletion | Chr10 | 8508332 | 8508341 | 9 | LOC_Os10g16990 |
| Deletion | Chr8 | 11148717 | 11148718 | 1 | LOC_Os08g18180 |
| Deletion | Chr12 | 14037047 | 14037048 | 1 | |
| Deletion | Chr6 | 14855080 | 14855092 | 12 | |
| Deletion | Chr6 | 14893001 | 14959000 | 66000 | 10 |
| Deletion | Chr2 | 15280600 | 15280606 | 6 | |
| Deletion | Chr8 | 16875679 | 16875682 | 3 | |
| Deletion | Chr8 | 17304032 | 17304033 | 1 | |
| Deletion | Chr8 | 17533910 | 17533911 | 1 | |
| Deletion | Chr1 | 18465560 | 18465562 | 2 | |
| Deletion | Chr8 | 19211625 | 19211629 | 4 | |
| Deletion | Chr12 | 19962612 | 19962613 | 1 | |
| Deletion | Chr10 | 22585480 | 22585481 | 1 | |
| Deletion | Chr2 | 24199178 | 24199183 | 5 | |
| Deletion | Chr3 | 26027211 | 26027212 | 1 | |
| Deletion | Chr4 | 26121521 | 26121551 | 30 | |
| Deletion | Chr6 | 26483001 | 27360000 | 877000 | 133 |
| Deletion | Chr12 | 27025881 | 27025883 | 2 | 2 |
| Deletion | Chr5 | 27057815 | 27057830 | 15 | LOC_Os05g46740 |
| Deletion | Chr2 | 27846684 | 27846698 | 14 | |
| Deletion | Chr1 | 38280898 | 38280906 | 8 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 27393345 | 27393345 | 1 | |
| Insertion | Chr4 | 21130579 | 21130580 | 2 | |
| Insertion | Chr9 | 18029290 | 18029291 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 26483171 | 27507005 | LOC_Os06g43960 |
No Translocation
