Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1453-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1453-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 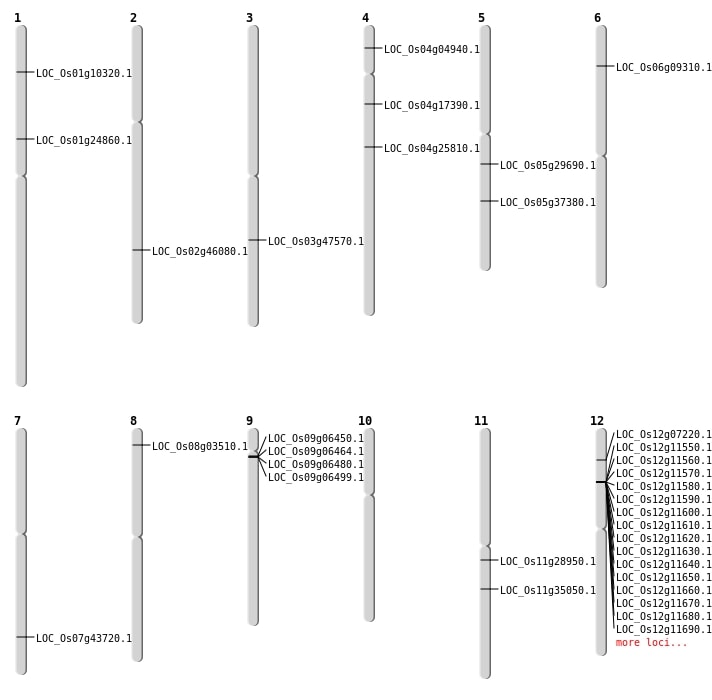
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20929093 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 40873408 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 13003509 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 5384461 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 20535821 | T-A | HOMO | STOP_GAINED | LOC_Os11g35050 |
| SBS | Chr11 | 704196 | A-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 13114755 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 14870079 | A-C | HET | UTR_5_PRIME | |
| SBS | Chr2 | 11456166 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 15820171 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 26900382 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g47570 |
| SBS | Chr4 | 17450760 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 30864825 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 32465768 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 3945472 | G-T | HET | INTRON | |
| SBS | Chr4 | 4524447 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 9516698 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g17390 |
| SBS | Chr5 | 17161912 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g29690 |
| SBS | Chr5 | 23544427 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 5689295 | G-A | HOMO | INTRON | |
| SBS | Chr5 | 8353111 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 19457744 | C-T | HET | INTRON | |
| SBS | Chr6 | 9047360 | C-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 16206027 | G-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 20775089 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 11707020 | T-A | HOMO | INTRON | |
| SBS | Chr8 | 19555953 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 20693617 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 3431886 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr8 | 5213599 | T-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 6586013 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 7055972 | A-T | HOMO | INTRON | |
| SBS | Chr8 | 7854250 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 655958 | C-T | HET | INTERGENIC |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 860017 | 860212 | 195 | |
| Deletion | Chr8 | 1642941 | 1642942 | 1 | LOC_Os08g03510 |
| Deletion | Chr7 | 2354095 | 2354125 | 30 | |
| Deletion | Chr4 | 2381584 | 2381604 | 20 | LOC_Os04g04940 |
| Deletion | Chr8 | 2474849 | 2474868 | 19 | |
| Deletion | Chr9 | 3035001 | 3086000 | 51000 | 4 |
| Deletion | Chr12 | 3552459 | 3552474 | 15 | LOC_Os12g07220 |
| Deletion | Chr1 | 3963075 | 3963080 | 5 | |
| Deletion | Chr11 | 4607464 | 4607468 | 4 | |
| Deletion | Chr6 | 4679019 | 4679030 | 11 | LOC_Os06g09310 |
| Deletion | Chr1 | 5429448 | 5429460 | 12 | LOC_Os01g10320 |
| Deletion | Chr12 | 6240379 | 6531897 | 291518 | 38 |
| Deletion | Chr9 | 6627660 | 6627664 | 4 | |
| Deletion | Chr7 | 7684423 | 7684438 | 15 | |
| Deletion | Chr4 | 9622560 | 9625360 | 2800 | |
| Deletion | Chr10 | 9766701 | 9766703 | 2 | |
| Deletion | Chr6 | 10725535 | 10725544 | 9 | |
| Deletion | Chr1 | 14001415 | 14001424 | 9 | LOC_Os01g24860 |
| Deletion | Chr3 | 14464197 | 14464200 | 3 | |
| Deletion | Chr4 | 14978433 | 14978435 | 2 | LOC_Os04g25810 |
| Deletion | Chr10 | 16122579 | 16122585 | 6 | |
| Deletion | Chr5 | 17022417 | 17022421 | 4 | |
| Deletion | Chr5 | 17477148 | 17477155 | 7 | |
| Deletion | Chr9 | 18372837 | 18372842 | 5 | |
| Deletion | Chr5 | 21870125 | 21870136 | 11 | LOC_Os05g37380 |
| Deletion | Chr8 | 22415153 | 22415154 | 1 | |
| Deletion | Chr8 | 24747742 | 24747757 | 15 | |
| Deletion | Chr12 | 25065615 | 25065631 | 16 | LOC_Os12g40510 |
| Deletion | Chr7 | 26163743 | 26163750 | 7 | LOC_Os07g43720 |
| Deletion | Chr11 | 27712164 | 27712174 | 10 | |
| Deletion | Chr5 | 28989315 | 28989317 | 2 | |
| Deletion | Chr1 | 30528066 | 30528076 | 10 | |
| Deletion | Chr1 | 30903405 | 30903441 | 36 | |
| Deletion | Chr3 | 35717958 | 35717986 | 28 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 14416901 | 14416901 | 1 | |
| Insertion | Chr12 | 1185381 | 1185381 | 1 | |
| Insertion | Chr2 | 28081723 | 28081724 | 2 | LOC_Os02g46080 |
| Insertion | Chr5 | 22811248 | 22811249 | 2 | |
| Insertion | Chr8 | 7934523 | 7934523 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 15463446 | 16754506 | LOC_Os11g28950 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 8525857 | Chr10 | 22976148 |
