Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1458-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1458-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 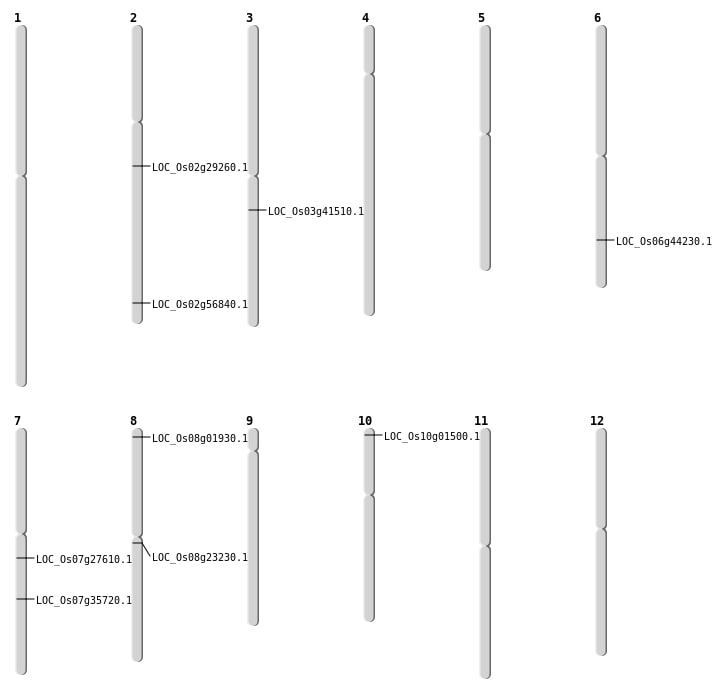
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 31704951 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 41827538 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 10845900 | C-T | HOMO | INTRON | |
| SBS | Chr11 | 15014208 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 5832180 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 14158650 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 17115324 | G-T | HET | INTRON | |
| SBS | Chr12 | 24376660 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 17648381 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 28735908 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 34835550 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g56840 |
| SBS | Chr2 | 34835551 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 7023269 | C-T | HET | INTRON | |
| SBS | Chr3 | 12583811 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 26112283 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 32249801 | G-C | HOMO | INTRON | |
| SBS | Chr4 | 10030843 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 12920960 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 13080978 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 1357859 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 17597885 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 17630114 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 18295699 | T-G | HET | INTRON | |
| SBS | Chr5 | 18470936 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 23970692 | C-T | HOMO | INTRON | |
| SBS | Chr5 | 24778346 | T-A | HOMO | INTRON | |
| SBS | Chr6 | 10914047 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 11383675 | T-A | HET | INTRON | |
| SBS | Chr6 | 17186426 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 2792366 | A-T | HOMO | INTRON | |
| SBS | Chr6 | 28261903 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 16129706 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g27610 |
| SBS | Chr7 | 16143620 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 13668400 | G-C | HET | INTRON | |
| SBS | Chr8 | 14026605 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g23230 |
| SBS | Chr8 | 25006433 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 5160174 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 3237368 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 4209820 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 8331468 | G-A | HET | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 321345 | 321347 | 2 | LOC_Os10g01500 |
| Deletion | Chr8 | 577757 | 577760 | 3 | LOC_Os08g01930 |
| Deletion | Chr11 | 2819922 | 2819938 | 16 | |
| Deletion | Chr3 | 4421687 | 4421688 | 1 | |
| Deletion | Chr2 | 5215279 | 5215289 | 10 | |
| Deletion | Chr9 | 6206546 | 6206554 | 8 | |
| Deletion | Chr5 | 6392116 | 6392128 | 12 | |
| Deletion | Chr1 | 9873867 | 9873868 | 1 | |
| Deletion | Chr2 | 11094872 | 11094873 | 1 | |
| Deletion | Chr6 | 11352627 | 11352629 | 2 | |
| Deletion | Chr5 | 15202774 | 15202781 | 7 | |
| Deletion | Chr2 | 17387178 | 17387184 | 6 | LOC_Os02g29260 |
| Deletion | Chr7 | 21392609 | 21392610 | 1 | LOC_Os07g35720 |
| Deletion | Chr8 | 21440466 | 21440470 | 4 | |
| Deletion | Chr11 | 22359293 | 22359302 | 9 | |
| Deletion | Chr3 | 23088049 | 23092741 | 4692 | LOC_Os03g41510 |
| Deletion | Chr12 | 24398534 | 24398538 | 4 | |
| Deletion | Chr5 | 26568155 | 26568156 | 1 | |
| Deletion | Chr8 | 27014819 | 27014821 | 2 |
Insertions: 10
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 26406024 | 26686431 | LOC_Os06g44230 |
| Inversion | Chr6 | 26406036 | 26686428 | LOC_Os06g44230 |
No Translocation
