Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1462-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1462-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 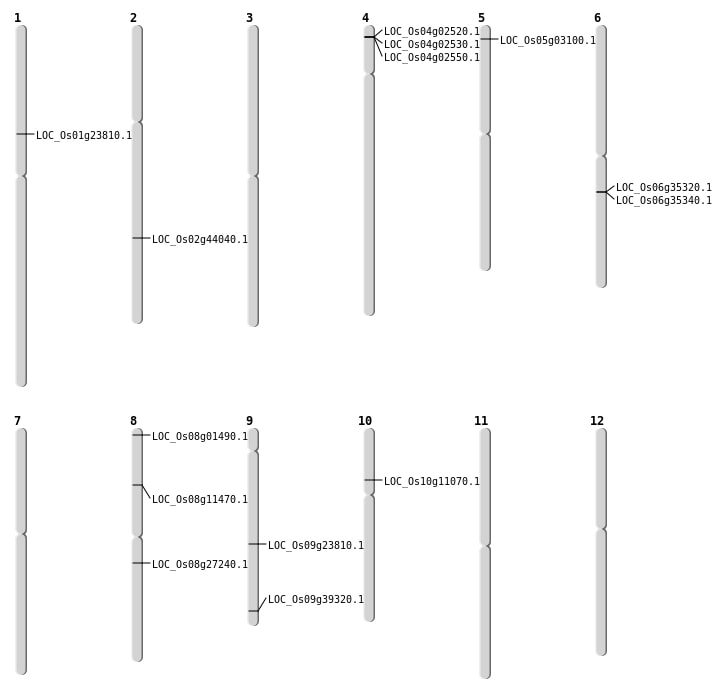
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21309711 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 4683527 | C-T | HET | INTRON | |
| SBS | Chr10 | 5128295 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 5898168 | G-T | Homo | INTERGENIC | |
| SBS | Chr11 | 13886395 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 1155717 | C-A | HET | UTR_5_PRIME | |
| SBS | Chr12 | 23994511 | C-T | HET | INTRON | |
| SBS | Chr12 | 25173153 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 8610822 | G-T | Homo | INTRON | |
| SBS | Chr2 | 26586538 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g44040 |
| SBS | Chr2 | 5564020 | A-G | Homo | INTERGENIC | |
| SBS | Chr3 | 14150061 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr3 | 14150062 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr3 | 20125629 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 487107 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 8450083 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 8408192 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 1228267 | A-T | HET | STOP_GAINED | LOC_Os05g03100 |
| SBS | Chr5 | 29889987 | C-T | HET | INTRON | |
| SBS | Chr6 | 30436203 | G-A | Homo | INTRON | |
| SBS | Chr6 | 9778746 | C-T | HET | INTRON | |
| SBS | Chr7 | 20783800 | G-A | Homo | INTRON | |
| SBS | Chr7 | 21018595 | G-A | Homo | INTERGENIC | |
| SBS | Chr7 | 21855712 | T-A | Homo | INTRON | |
| SBS | Chr8 | 1135130 | G-A | HET | INTRON | |
| SBS | Chr8 | 13777220 | G-A | Homo | SYNONYMOUS_CODING | |
| SBS | Chr8 | 23244788 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 28107109 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 6730861 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g11470 |
| SBS | Chr9 | 14189122 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g23810 |
Deletions: 39
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 5970 | 5976 | 6 | |
| Deletion | Chr8 | 302374 | 302375 | 1 | LOC_Os08g01490 |
| Deletion | Chr1 | 550853 | 550865 | 12 | |
| Deletion | Chr8 | 833101 | 833102 | 1 | |
| Deletion | Chr4 | 918001 | 948000 | 30000 | 3 |
| Deletion | Chr4 | 1749448 | 1749450 | 2 | |
| Deletion | Chr4 | 2398790 | 2398791 | 1 | |
| Deletion | Chr7 | 3411009 | 3411027 | 18 | |
| Deletion | Chr10 | 4644970 | 4644971 | 1 | |
| Deletion | Chr8 | 5102283 | 5102808 | 525 | |
| Deletion | Chr9 | 5446363 | 5446366 | 3 | |
| Deletion | Chr10 | 6122942 | 6122945 | 3 | LOC_Os10g11070 |
| Deletion | Chr8 | 8151945 | 8151950 | 5 | |
| Deletion | Chr6 | 9778727 | 9778736 | 9 | |
| Deletion | Chr3 | 10228445 | 10228491 | 46 | |
| Deletion | Chr6 | 11331163 | 11331213 | 50 | |
| Deletion | Chr6 | 12208940 | 12208941 | 1 | |
| Deletion | Chr8 | 12963897 | 12963902 | 5 | |
| Deletion | Chr1 | 13396355 | 13396357 | 2 | LOC_Os01g23810 |
| Deletion | Chr3 | 14100458 | 14100459 | 1 | |
| Deletion | Chr6 | 14126579 | 14126580 | 1 | |
| Deletion | Chr1 | 14195946 | 14195951 | 5 | |
| Deletion | Chr1 | 16839712 | 16839713 | 1 | |
| Deletion | Chr6 | 20247483 | 20247520 | 37 | |
| Deletion | Chr6 | 20586603 | 20604297 | 17694 | 2 |
| Deletion | Chr8 | 21108492 | 21108493 | 1 | |
| Deletion | Chr4 | 22116606 | 22116631 | 25 | |
| Deletion | Chr9 | 22604614 | 22604626 | 12 | LOC_Os09g39320 |
| Deletion | Chr6 | 24235698 | 24235706 | 8 | |
| Deletion | Chr1 | 24704067 | 24704068 | 1 | |
| Deletion | Chr12 | 25633644 | 25633647 | 3 | |
| Deletion | Chr3 | 29492827 | 29492830 | 3 | |
| Deletion | Chr4 | 30061576 | 30061577 | 1 | |
| Deletion | Chr3 | 30314812 | 30314912 | 100 | |
| Deletion | Chr1 | 31178619 | 31178628 | 9 | |
| Deletion | Chr3 | 31779351 | 31779352 | 1 | |
| Deletion | Chr1 | 32397393 | 32397394 | 1 | |
| Deletion | Chr1 | 35671123 | 35671125 | 2 | |
| Deletion | Chr1 | 42382129 | 42382133 | 4 |
Insertions: 6
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 11420162 | 16628348 | LOC_Os08g27240 |
| Inversion | Chr8 | 11420173 | 16628354 | LOC_Os08g27240 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 9583663 | Chr3 | 30314912 |
