Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1467-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1467-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 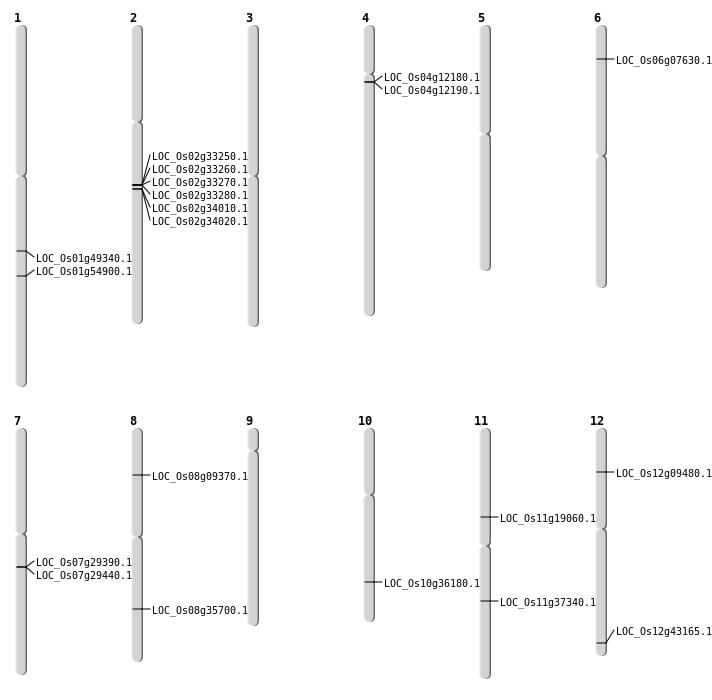
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1346163 | T-A | HET | UTR_5_PRIME | |
| SBS | Chr1 | 30424695 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 35831596 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 39921292 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 19325111 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 19325112 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 9746421 | A-T | HET | INTRON | |
| SBS | Chr11 | 10865951 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g19060 |
| SBS | Chr11 | 25832413 | C-T | HET | INTRON | |
| SBS | Chr12 | 19663106 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 26806715 | A-G | HOMO | START_GAINED | LOC_Os12g43165 |
| SBS | Chr12 | 2749946 | G-T | HET | INTRON | |
| SBS | Chr12 | 3292235 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 4972251 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g09480 |
| SBS | Chr2 | 21878221 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 21878222 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 21878224 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 7267084 | A-T | HOMO | UTR_3_PRIME | |
| SBS | Chr3 | 18714187 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 33419427 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 34355514 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 34400916 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 22642831 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 28317851 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 3063402 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 33511067 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 692542 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 8397858 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 8888993 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 26093717 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 29366762 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 3292372 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 3692102 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g07630 |
| SBS | Chr7 | 10540604 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 2454744 | A-G | HOMO | INTRON | |
| SBS | Chr7 | 28828224 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 22527376 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g35700 |
| SBS | Chr8 | 22892709 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr8 | 25256038 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 26083689 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 26083716 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 14624644 | A-G | HET | INTERGENIC |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 1453901 | 1453907 | 6 | |
| Deletion | Chr10 | 7525623 | 7525654 | 31 | |
| Deletion | Chr10 | 11209388 | 11209389 | 1 | |
| Deletion | Chr3 | 14361116 | 14361130 | 14 | |
| Deletion | Chr10 | 16891561 | 16891562 | 1 | |
| Deletion | Chr7 | 17276230 | 17276233 | 3 | LOC_Os07g29440 |
| Deletion | Chr3 | 17779812 | 17779837 | 25 | |
| Deletion | Chr2 | 19761039 | 19780526 | 19487 | 4 |
| Deletion | Chr2 | 20302958 | 20320462 | 17504 | 2 |
| Deletion | Chr11 | 22045801 | 22045802 | 1 | LOC_Os11g37340 |
| Deletion | Chr4 | 22319899 | 22319901 | 2 | |
| Deletion | Chr3 | 25652515 | 25652516 | 1 | |
| Deletion | Chr4 | 27001544 | 27001561 | 17 | |
| Deletion | Chr1 | 28368320 | 28368334 | 14 | LOC_Os01g49340 |
| Deletion | Chr3 | 33407042 | 33407047 | 5 | |
| Deletion | Chr3 | 34444783 | 34444795 | 12 |
Insertions: 5
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 6710805 | 7764206 | LOC_Os04g12180 |
| Inversion | Chr4 | 6715015 | 7764213 | LOC_Os04g12190 |
| Inversion | Chr10 | 19327190 | 19347194 | LOC_Os10g36180 |
| Inversion | Chr1 | 31569469 | 31756576 | LOC_Os01g54900 |
| Inversion | Chr1 | 31569741 | 31756584 | LOC_Os01g54900 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr2 | 4169616 | Chr1 | 31569477 | 2 |
| Translocation | Chr8 | 5063837 | Chr6 | 3022425 | |
| Translocation | Chr8 | 5435246 | Chr2 | 1621705 | LOC_Os02g03840 |
| Translocation | Chr8 | 5436163 | Chr1 | 31569731 | 2 |
| Translocation | Chr8 | 5436210 | Chr2 | 4169667 | 2 |
| Translocation | Chr4 | 10240756 | Chr2 | 26403808 | |
| Translocation | Chr7 | 17257745 | Chr1 | 29500429 | LOC_Os07g29390 |
| Translocation | Chr7 | 17257746 | Chr1 | 29500430 | LOC_Os07g29390 |
