Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1468-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1468-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 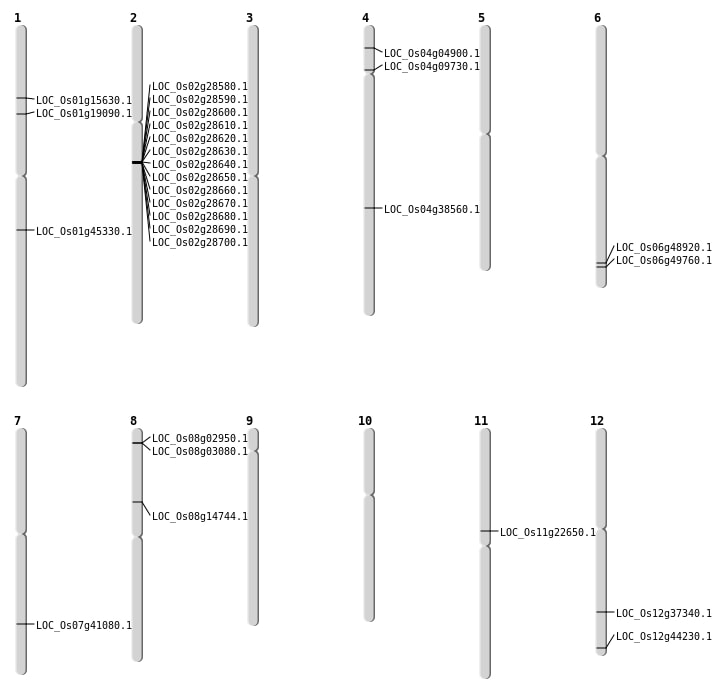
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22090314 | C-A | HET | INTRON | |
| SBS | Chr1 | 24512984 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 25732281 | G-A | HET | STOP_GAINED | LOC_Os01g45330 |
| SBS | Chr1 | 33087102 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 38742937 | T-C | HET | INTRON | |
| SBS | Chr1 | 39098066 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 14018423 | T-A | Homo | INTERGENIC | |
| SBS | Chr11 | 9135681 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 15391786 | A-G | HET | INTRON | |
| SBS | Chr12 | 22929920 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g37340 |
| SBS | Chr2 | 12834194 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 12834195 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 16607962 | C-G | HET | INTRON | |
| SBS | Chr3 | 1003855 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 27694354 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 3142692 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 10465734 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 22911599 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g38560 |
| SBS | Chr4 | 5222379 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g09730 |
| SBS | Chr6 | 2617627 | C-T | Homo | INTRON | |
| SBS | Chr7 | 9343932 | C-T | Homo | INTERGENIC | |
| SBS | Chr8 | 12488994 | A-C | Homo | SYNONYMOUS_CODING | |
| SBS | Chr8 | 17475520 | A-T | HET | INTRON | |
| SBS | Chr8 | 8870677 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g14744 |
| SBS | Chr9 | 11041965 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 15750896 | G-A | Homo | INTERGENIC | |
| SBS | Chr9 | 4018404 | A-C | HET | INTRON |
Deletions: 38
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 450725 | 450757 | 32 | |
| Deletion | Chr2 | 1334150 | 1334160 | 10 | |
| Deletion | Chr4 | 2362749 | 2362762 | 13 | LOC_Os04g04900 |
| Deletion | Chr4 | 2907856 | 2907857 | 1 | |
| Deletion | Chr5 | 4444731 | 4444733 | 2 | |
| Deletion | Chr11 | 4922739 | 4922740 | 1 | |
| Deletion | Chr12 | 5500830 | 5500838 | 8 | |
| Deletion | Chr1 | 5604794 | 5604807 | 13 | |
| Deletion | Chr4 | 6640200 | 6640206 | 6 | |
| Deletion | Chr8 | 6841552 | 6841553 | 1 | |
| Deletion | Chr7 | 7216298 | 7216304 | 6 | |
| Deletion | Chr1 | 8790922 | 8790924 | 2 | LOC_Os01g15630 |
| Deletion | Chr7 | 9559331 | 9559341 | 10 | |
| Deletion | Chr6 | 9594968 | 9594970 | 2 | |
| Deletion | Chr2 | 10187948 | 10187953 | 5 | |
| Deletion | Chr12 | 10618566 | 10618568 | 2 | |
| Deletion | Chr1 | 10788296 | 10788299 | 3 | LOC_Os01g19090 |
| Deletion | Chr2 | 12973932 | 12973934 | 2 | |
| Deletion | Chr11 | 13030898 | 13033251 | 2353 | LOC_Os11g22650 |
| Deletion | Chr6 | 14934626 | 14934627 | 1 | |
| Deletion | Chr2 | 16916001 | 16980000 | 64000 | 13 |
| Deletion | Chr12 | 17634801 | 17634803 | 2 | |
| Deletion | Chr5 | 18078740 | 18078745 | 5 | |
| Deletion | Chr12 | 19591770 | 19591806 | 36 | |
| Deletion | Chr2 | 20227852 | 20227871 | 19 | |
| Deletion | Chr8 | 20486091 | 20486095 | 4 | |
| Deletion | Chr1 | 21956832 | 21956833 | 1 | |
| Deletion | Chr1 | 22278311 | 22278316 | 5 | |
| Deletion | Chr10 | 22395409 | 22395425 | 16 | |
| Deletion | Chr8 | 23588689 | 23588693 | 4 | |
| Deletion | Chr2 | 24898699 | 24898711 | 12 | |
| Deletion | Chr12 | 27430366 | 27430370 | 4 | LOC_Os12g44230 |
| Deletion | Chr6 | 28744622 | 28744623 | 1 | |
| Deletion | Chr4 | 29123347 | 29123361 | 14 | |
| Deletion | Chr6 | 30106588 | 30106589 | 1 | LOC_Os06g49760 |
| Deletion | Chr1 | 31865077 | 31865078 | 1 | |
| Deletion | Chr1 | 35434708 | 35434709 | 1 | |
| Deletion | Chr1 | 41773918 | 41773923 | 5 |
Insertions: 18
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 648329 | 24575314 | LOC_Os07g41080 |
| Inversion | Chr8 | 1283774 | 1381267 | 2 |
| Inversion | Chr8 | 1283801 | 1382306 | 2 |
| Inversion | Chr7 | 24573899 | 24574263 | |
| Inversion | Chr6 | 28985278 | 29616941 | LOC_Os06g48920 |
No Translocation
