Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1473-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1473-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 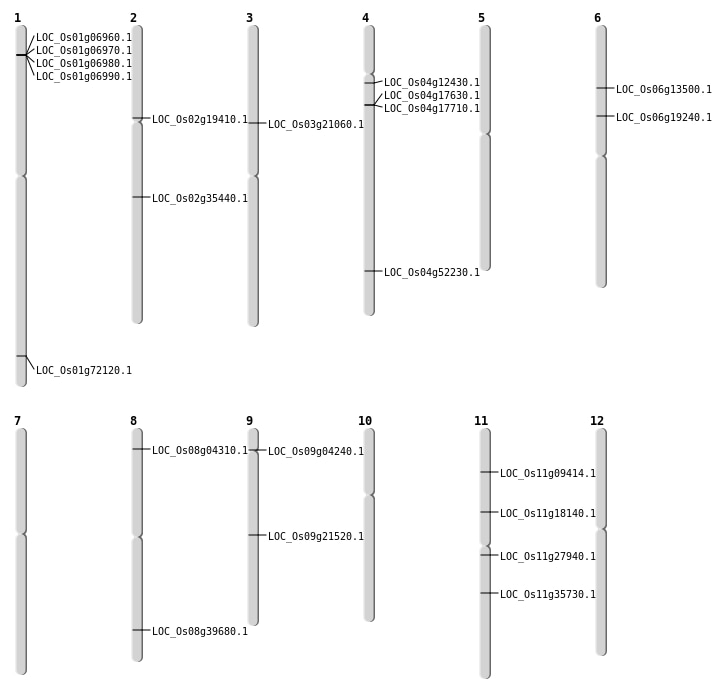
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16767809 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 41841047 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g72120 |
| SBS | Chr1 | 9909848 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 7214163 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 8633438 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 14480752 | G-A | Homo | INTRON | |
| SBS | Chr11 | 16074557 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g27940 |
| SBS | Chr11 | 18344360 | C-A | Homo | INTERGENIC | |
| SBS | Chr11 | 8088195 | A-G | Homo | INTERGENIC | |
| SBS | Chr2 | 10199560 | T-A | HET | INTRON | |
| SBS | Chr2 | 27042944 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 7739451 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 13261868 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 23649278 | G-A | HET | INTRON | |
| SBS | Chr3 | 2997285 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 23283993 | A-G | Homo | INTERGENIC | |
| SBS | Chr4 | 32928482 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 23022831 | A-T | Homo | INTERGENIC | |
| SBS | Chr6 | 1146684 | G-A | Homo | INTERGENIC | |
| SBS | Chr6 | 15028041 | G-A | HET | INTRON | |
| SBS | Chr6 | 15572995 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 25869104 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 7443142 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g13500 |
| SBS | Chr7 | 14954331 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 21992843 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 9560290 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 21747708 | T-A | Homo | INTERGENIC | |
| SBS | Chr8 | 6777746 | C-G | Homo | INTRON | |
| SBS | Chr9 | 14445008 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 15683814 | A-G | Homo | INTERGENIC |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 261925 | 261933 | 8 | |
| Deletion | Chr1 | 1668713 | 1668721 | 8 | |
| Deletion | Chr9 | 2244135 | 2244144 | 9 | LOC_Os09g04240 |
| Deletion | Chr1 | 3291119 | 3298000 | 6881 | 4 |
| Deletion | Chr1 | 3390881 | 3390933 | 52 | |
| Deletion | Chr9 | 3566760 | 3566772 | 12 | |
| Deletion | Chr4 | 5166405 | 5166415 | 10 | |
| Deletion | Chr7 | 6145047 | 6145077 | 30 | |
| Deletion | Chr4 | 7587336 | 7587338 | 2 | |
| Deletion | Chr11 | 8839144 | 8839146 | 2 | |
| Deletion | Chr6 | 9136839 | 9136857 | 18 | |
| Deletion | Chr11 | 10208776 | 10208777 | 1 | LOC_Os11g18140 |
| Deletion | Chr6 | 10948621 | 10948622 | 1 | LOC_Os06g19240 |
| Deletion | Chr9 | 10967538 | 10967555 | 17 | |
| Deletion | Chr3 | 11974482 | 11974489 | 7 | LOC_Os03g21060 |
| Deletion | Chr9 | 13019868 | 13021627 | 1759 | LOC_Os09g21520 |
| Deletion | Chr9 | 13723122 | 13723152 | 30 | |
| Deletion | Chr10 | 14720405 | 14720427 | 22 | |
| Deletion | Chr3 | 15774714 | 15774715 | 1 | |
| Deletion | Chr1 | 17330029 | 17330030 | 1 | |
| Deletion | Chr12 | 19333288 | 19333299 | 11 | |
| Deletion | Chr3 | 19944688 | 19944689 | 1 | |
| Deletion | Chr11 | 20063236 | 20063238 | 2 | |
| Deletion | Chr5 | 20439360 | 20439361 | 1 | |
| Deletion | Chr11 | 20976959 | 20976960 | 1 | LOC_Os11g35730 |
| Deletion | Chr12 | 21051656 | 21051657 | 1 | |
| Deletion | Chr8 | 21063090 | 21063091 | 1 | |
| Deletion | Chr4 | 21274688 | 21274689 | 1 | |
| Deletion | Chr2 | 21304549 | 21304558 | 9 | LOC_Os02g35440 |
| Deletion | Chr8 | 22733661 | 22733670 | 9 | |
| Deletion | Chr8 | 25116211 | 25116220 | 9 | LOC_Os08g39680 |
| Deletion | Chr2 | 27654437 | 27654455 | 18 |
Insertions: 9
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 10105196 | 10105196 | 1 | |
| Insertion | Chr1 | 19910040 | 19910040 | 1 | |
| Insertion | Chr11 | 5051071 | 5051071 | 1 | LOC_Os11g09414 |
| Insertion | Chr12 | 11007901 | 11007902 | 2 | |
| Insertion | Chr2 | 11335492 | 11335494 | 3 | LOC_Os02g19410 |
| Insertion | Chr2 | 24309576 | 24309577 | 2 | |
| Insertion | Chr4 | 31026858 | 31026901 | 44 | LOC_Os04g52230 |
| Insertion | Chr6 | 7561931 | 7561932 | 2 | |
| Insertion | Chr8 | 2115886 | 2115895 | 10 | LOC_Os08g04310 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 6870287 | 9704732 | 2 |
| Inversion | Chr4 | 6870602 | 9649970 | 2 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 8141544 | Chr4 | 6870297 | LOC_Os04g12430 |
| Translocation | Chr11 | 8141608 | Chr4 | 6870630 | LOC_Os04g12430 |
| Translocation | Chr6 | 14438352 | Chr4 | 9649194 | LOC_Os04g17630 |
