Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1475-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1475-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 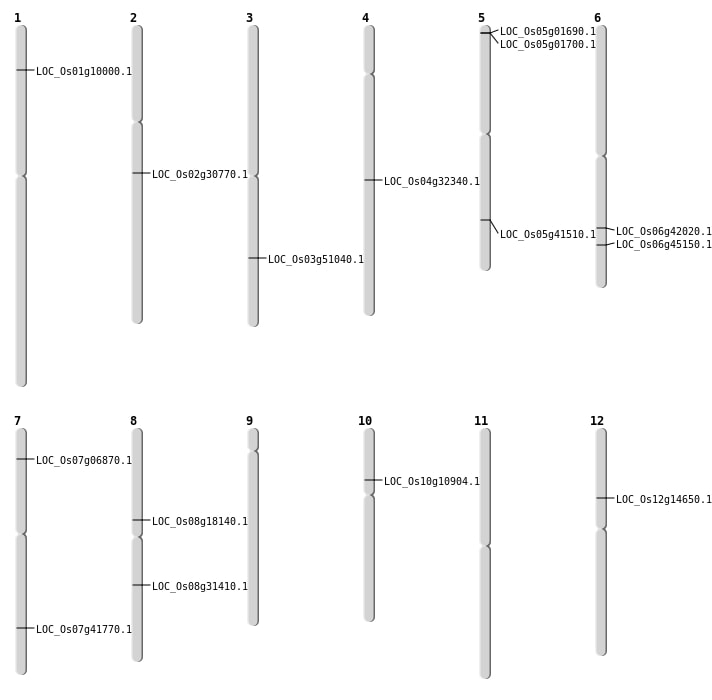
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12230617 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 14165816 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 31016139 | A-T | Homo | INTRON | |
| SBS | Chr1 | 31033721 | T-G | Homo | INTERGENIC | |
| SBS | Chr1 | 38881855 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 11482731 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 20628203 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 8377680 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g14650 |
| SBS | Chr12 | 9457353 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 30007821 | G-A | Homo | INTERGENIC | |
| SBS | Chr2 | 5527303 | A-C | HET | INTRON | |
| SBS | Chr3 | 14454112 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 17229709 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 20359367 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 21426818 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 2894154 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 29183105 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g51040 |
| SBS | Chr4 | 19896923 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 12755888 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 18902586 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 24307624 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g41510 |
| SBS | Chr5 | 5612458 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 604451 | G-T | Homo | INTERGENIC | |
| SBS | Chr6 | 25231596 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g42020 |
| SBS | Chr7 | 10830532 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 11297541 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 12614806 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 2466085 | A-C | HET | INTRON | |
| SBS | Chr7 | 26244104 | C-T | HET | INTRON | |
| SBS | Chr7 | 5070662 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 19429395 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g31410 |
| SBS | Chr9 | 11446740 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 12509300 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 13401876 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 14818343 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 17900087 | A-T | Homo | INTERGENIC | |
| SBS | Chr9 | 17900088 | C-G | Homo | INTERGENIC | |
| SBS | Chr9 | 6880689 | T-A | Homo | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 688333 | 688334 | 1 | |
| Deletion | Chr10 | 3848072 | 3848074 | 2 | |
| Deletion | Chr11 | 4732980 | 4732988 | 8 | |
| Deletion | Chr10 | 4871521 | 4871523 | 2 | |
| Deletion | Chr10 | 6047376 | 6047397 | 21 | LOC_Os10g10904 |
| Deletion | Chr5 | 8336631 | 8336633 | 2 | |
| Deletion | Chr3 | 13864819 | 13864821 | 2 | |
| Deletion | Chr9 | 14429195 | 14429205 | 10 | |
| Deletion | Chr8 | 14433740 | 14433742 | 2 | |
| Deletion | Chr9 | 14637146 | 14637153 | 7 | |
| Deletion | Chr4 | 16254513 | 16254521 | 8 | |
| Deletion | Chr12 | 16487803 | 16487809 | 6 | |
| Deletion | Chr12 | 17272847 | 17272849 | 2 | |
| Deletion | Chr11 | 19282698 | 19282699 | 1 | |
| Deletion | Chr2 | 20497641 | 20497658 | 17 | |
| Deletion | Chr1 | 21045122 | 21045124 | 2 | |
| Deletion | Chr9 | 21717301 | 21717314 | 13 | |
| Deletion | Chr3 | 21748454 | 21748455 | 1 | |
| Deletion | Chr5 | 22830001 | 22830002 | 1 | |
| Deletion | Chr2 | 24022495 | 24022503 | 8 | |
| Deletion | Chr7 | 24599852 | 24599853 | 1 | |
| Deletion | Chr7 | 25025065 | 25025066 | 1 | LOC_Os07g41770 |
| Deletion | Chr6 | 27303608 | 27303609 | 1 | LOC_Os06g45150 |
| Deletion | Chr5 | 28114197 | 28114231 | 34 | |
| Deletion | Chr7 | 28279984 | 28279988 | 4 | |
| Deletion | Chr1 | 29597119 | 29597120 | 1 | |
| Deletion | Chr4 | 29664069 | 29664070 | 1 | |
| Deletion | Chr5 | 29796324 | 29796325 | 1 |
Insertions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 22029103 | 22029116 | 14 | |
| Insertion | Chr10 | 17413274 | 17413297 | 24 | |
| Insertion | Chr10 | 4299561 | 4299590 | 30 | |
| Insertion | Chr12 | 1015859 | 1015879 | 21 | |
| Insertion | Chr2 | 18330104 | 18330127 | 24 | LOC_Os02g30770 |
| Insertion | Chr4 | 7017770 | 7017771 | 2 | |
| Insertion | Chr5 | 18483723 | 18483723 | 1 | |
| Insertion | Chr5 | 8560911 | 8560911 | 1 | |
| Insertion | Chr6 | 10216060 | 10216060 | 1 | |
| Insertion | Chr6 | 18663246 | 18663246 | 1 | |
| Insertion | Chr7 | 26244093 | 26244094 | 2 | |
| Insertion | Chr7 | 3359447 | 3359456 | 10 | LOC_Os07g06870 |
| Insertion | Chr8 | 11392455 | 11392455 | 1 | |
| Insertion | Chr8 | 6080851 | 6080851 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 5211832 | 5310504 | 2 |
| Inversion | Chr1 | 5212899 | 5310515 | 2 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 412478 | Chr4 | 19388011 | 2 |
| Translocation | Chr5 | 425588 | Chr4 | 19387892 | 2 |
| Translocation | Chr8 | 11122928 | Chr3 | 20627755 | LOC_Os08g18140 |
| Translocation | Chr8 | 18937202 | Chr1 | 5212876 | LOC_Os01g10000 |
| Translocation | Chr8 | 19401010 | Chr1 | 5211846 | LOC_Os01g10000 |
