Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1478-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1478-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 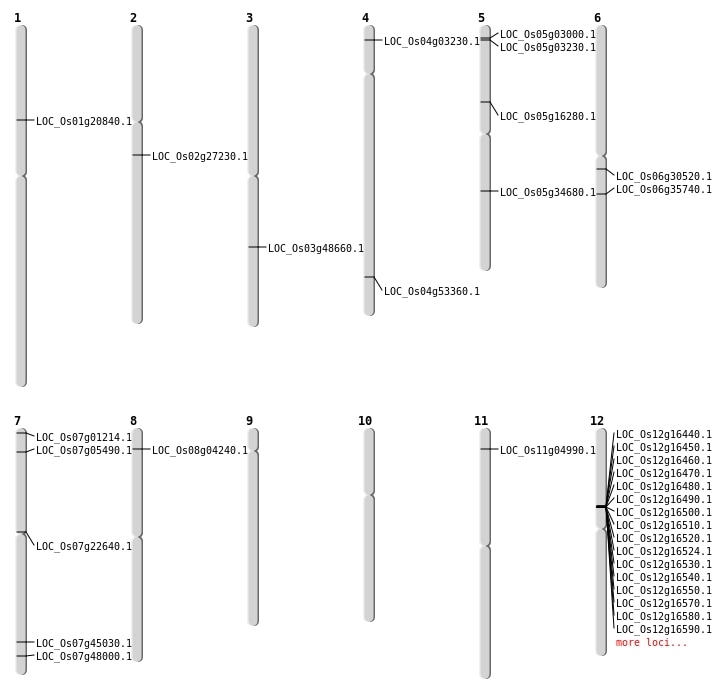
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 35322221 | T-C | HET | INTRON | |
| SBS | Chr1 | 39539917 | C-T | Homo | INTRON | |
| SBS | Chr1 | 7845313 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 8030658 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 8305887 | C-T | Homo | UTR_3_PRIME | |
| SBS | Chr10 | 327999 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 9333840 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 1529355 | A-T | HET | INTRON | |
| SBS | Chr12 | 19927565 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 1211243 | T-C | Homo | INTERGENIC | |
| SBS | Chr2 | 16025743 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g27230 |
| SBS | Chr3 | 12355782 | G-A | HET | INTRON | |
| SBS | Chr3 | 17397589 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 13700494 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 3947226 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 15711293 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 16822508 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 18638646 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 23723271 | G-A | HET | INTRON | |
| SBS | Chr6 | 1107683 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 16123780 | G-A | Homo | INTERGENIC | |
| SBS | Chr6 | 16306382 | G-T | Homo | INTERGENIC | |
| SBS | Chr6 | 17656121 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g30520 |
| SBS | Chr6 | 22422828 | C-T | Homo | INTERGENIC | |
| SBS | Chr6 | 24911544 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 4198673 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 5489616 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 2024457 | G-T | HET | INTRON | |
| SBS | Chr7 | 26871529 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g45030 |
| SBS | Chr8 | 11991730 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 27921511 | G-T | Homo | INTRON | |
| SBS | Chr8 | 6685630 | T-C | HET | INTERGENIC |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 122496 | 122505 | 9 | LOC_Os07g01214 |
| Deletion | Chr6 | 316828 | 316844 | 16 | |
| Deletion | Chr5 | 1130222 | 1130228 | 6 | LOC_Os05g03000 |
| Deletion | Chr5 | 1308231 | 1308233 | 2 | LOC_Os05g03230 |
| Deletion | Chr7 | 2562317 | 2562318 | 1 | LOC_Os07g05490 |
| Deletion | Chr8 | 3586676 | 3586677 | 1 | |
| Deletion | Chr10 | 4855331 | 4855332 | 1 | |
| Deletion | Chr11 | 5714599 | 5714601 | 2 | |
| Deletion | Chr11 | 5785664 | 5785665 | 1 | |
| Deletion | Chr5 | 6539473 | 6539474 | 1 | |
| Deletion | Chr12 | 6579285 | 6579286 | 1 | |
| Deletion | Chr6 | 6645585 | 6645587 | 2 | |
| Deletion | Chr9 | 6868776 | 6868777 | 1 | |
| Deletion | Chr8 | 8388697 | 8388699 | 2 | |
| Deletion | Chr10 | 9069847 | 9069852 | 5 | |
| Deletion | Chr5 | 9222825 | 9222826 | 1 | LOC_Os05g16280 |
| Deletion | Chr6 | 10263451 | 10263452 | 1 | |
| Deletion | Chr9 | 12002210 | 12002250 | 40 | |
| Deletion | Chr4 | 13970121 | 13970122 | 1 | |
| Deletion | Chr7 | 14428825 | 14428831 | 6 | |
| Deletion | Chr7 | 15569822 | 15569833 | 11 | |
| Deletion | Chr9 | 15981203 | 15981204 | 1 | |
| Deletion | Chr8 | 18535036 | 18535062 | 26 | |
| Deletion | Chr4 | 19818159 | 19818161 | 2 | |
| Deletion | Chr6 | 20553686 | 20553687 | 1 | |
| Deletion | Chr5 | 20583019 | 20583020 | 1 | LOC_Os05g34680 |
| Deletion | Chr7 | 20918402 | 20918403 | 1 | |
| Deletion | Chr10 | 22744208 | 22744209 | 1 | |
| Deletion | Chr4 | 22859680 | 22859681 | 1 | |
| Deletion | Chr7 | 23329227 | 23329241 | 14 | |
| Deletion | Chr6 | 24848222 | 24848226 | 4 | |
| Deletion | Chr6 | 27481958 | 27481997 | 39 | |
| Deletion | Chr11 | 27774733 | 27774735 | 2 | |
| Deletion | Chr7 | 28667755 | 28667756 | 1 | LOC_Os07g48000 |
| Deletion | Chr5 | 29396564 | 29396568 | 4 | |
| Deletion | Chr4 | 31777671 | 31777673 | 2 | LOC_Os04g53360 |
| Deletion | Chr1 | 38794355 | 38794379 | 24 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 16649328 | 16649343 | 16 | |
| Insertion | Chr10 | 5332346 | 5332347 | 2 | |
| Insertion | Chr11 | 16262716 | 16262716 | 1 | |
| Insertion | Chr11 | 6813044 | 6813044 | 1 | |
| Insertion | Chr3 | 1999064 | 1999065 | 2 | |
| Insertion | Chr4 | 2359536 | 2359537 | 2 | |
| Insertion | Chr4 | 26443891 | 26443892 | 2 | |
| Insertion | Chr6 | 20860801 | 20860803 | 3 | LOC_Os06g35740 |
| Insertion | Chr6 | 5419944 | 5419944 | 1 | |
| Insertion | Chr6 | 8803213 | 8803213 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 1169406 | 1372477 | LOC_Os04g03230 |
| Inversion | Chr12 | 9522960 | 10254739 | LOC_Os12g17880 |
| Inversion | Chr12 | 9522971 | 9606766 | |
| Inversion | Chr7 | 12759632 | 12792462 | LOC_Os07g22640 |
| Inversion | Chr7 | 12759655 | 12792467 | LOC_Os07g22640 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 2147809 | Chr8 | 2064292 | 2 |
| Translocation | Chr11 | 2148049 | Chr8 | 2064210 | 2 |
| Translocation | Chr3 | 27738919 | Chr1 | 11612766 | LOC_Os03g48660 |
| Translocation | Chr3 | 27738924 | Chr1 | 11620913 | 2 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr12 | 9414259 | 9712973 | 298714 | 50 |
