Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1484-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1484-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 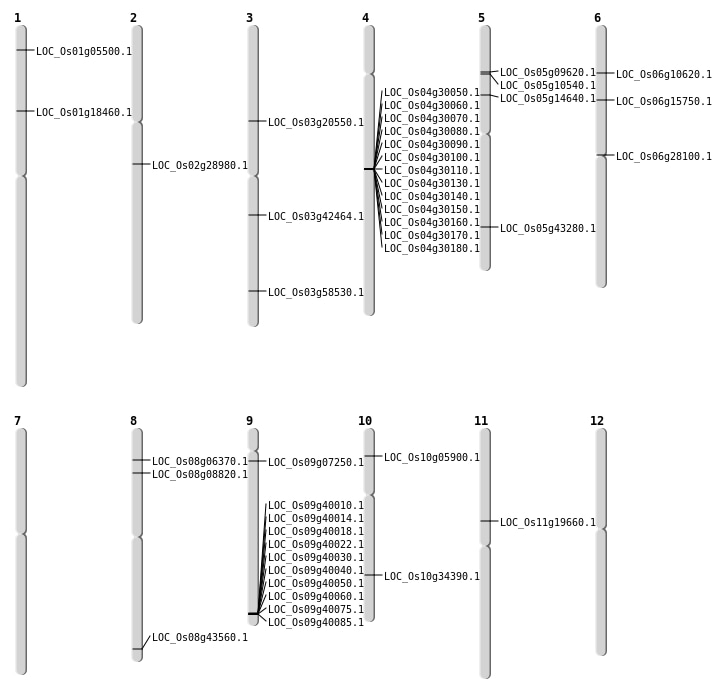
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13642546 | T-C | Homo | INTERGENIC | |
| SBS | Chr1 | 25374124 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 35752918 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 37564965 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 18365641 | G-T | HET | INTRON | |
| SBS | Chr10 | 18365643 | A-T | HET | INTRON | |
| SBS | Chr10 | 6353710 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 11327156 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g19660 |
| SBS | Chr11 | 1144673 | A-T | Homo | INTERGENIC | |
| SBS | Chr11 | 15996314 | A-T | Homo | INTERGENIC | |
| SBS | Chr11 | 15996353 | A-G | Homo | INTERGENIC | |
| SBS | Chr11 | 24811165 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 24242495 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 14609800 | G-A | Homo | INTERGENIC | |
| SBS | Chr2 | 1537019 | C-T | Homo | INTERGENIC | |
| SBS | Chr2 | 23145388 | A-G | Homo | INTERGENIC | |
| SBS | Chr2 | 23415048 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 29444521 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 13563957 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 7213140 | G-A | HET | INTRON | |
| SBS | Chr3 | 9863238 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 16596334 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 17227246 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 18596196 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 34736804 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 5733040 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g10540 |
| SBS | Chr5 | 681710 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 8332578 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g14640 |
| SBS | Chr5 | 8332579 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 28917145 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 474483 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 5524165 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g10620 |
| SBS | Chr6 | 8937053 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g15750 |
| SBS | Chr7 | 11442366 | T-C | Homo | INTERGENIC | |
| SBS | Chr7 | 13702283 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 17358067 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 18846251 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 20370270 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 25544614 | T-C | Homo | INTERGENIC | |
| SBS | Chr7 | 8097871 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 12081052 | T-C | HET | INTRON | |
| SBS | Chr9 | 3558922 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07250 |
| SBS | Chr9 | 5147195 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 9265043 | C-A | HET | UTR_5_PRIME |
Deletions: 38
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 983508 | 983510 | 2 | |
| Deletion | Chr1 | 2606462 | 2606543 | 81 | LOC_Os01g05500 |
| Deletion | Chr8 | 5033897 | 5033898 | 1 | |
| Deletion | Chr8 | 5116617 | 5116630 | 13 | LOC_Os08g08820 |
| Deletion | Chr5 | 5433045 | 5433052 | 7 | LOC_Os05g09620 |
| Deletion | Chr10 | 5709993 | 5709994 | 1 | |
| Deletion | Chr2 | 6427637 | 6427638 | 1 | |
| Deletion | Chr3 | 7363994 | 7364005 | 11 | |
| Deletion | Chr4 | 7839598 | 7839599 | 1 | |
| Deletion | Chr6 | 8481126 | 8481127 | 1 | |
| Deletion | Chr7 | 8885925 | 8885932 | 7 | |
| Deletion | Chr6 | 9648498 | 9648503 | 5 | |
| Deletion | Chr1 | 10376028 | 10376036 | 8 | LOC_Os01g18460 |
| Deletion | Chr5 | 11887886 | 11887893 | 7 | |
| Deletion | Chr6 | 12017739 | 12017740 | 1 | |
| Deletion | Chr4 | 12565220 | 12565222 | 2 | |
| Deletion | Chr5 | 13045556 | 13045557 | 1 | |
| Deletion | Chr4 | 14101714 | 14101730 | 16 | |
| Deletion | Chr2 | 14191910 | 14191911 | 1 | |
| Deletion | Chr6 | 15957654 | 15957655 | 1 | LOC_Os06g28100 |
| Deletion | Chr9 | 16376462 | 16376474 | 12 | |
| Deletion | Chr11 | 16563744 | 16563745 | 1 | |
| Deletion | Chr10 | 16997563 | 16997582 | 19 | |
| Deletion | Chr2 | 17158897 | 17158898 | 1 | LOC_Os02g28980 |
| Deletion | Chr4 | 17920001 | 18014000 | 94000 | 14 |
| Deletion | Chr10 | 18337046 | 18337055 | 9 | LOC_Os10g34390 |
| Deletion | Chr5 | 18469990 | 18470001 | 11 | |
| Deletion | Chr6 | 19914425 | 19914426 | 1 | |
| Deletion | Chr6 | 20028654 | 20028656 | 2 | |
| Deletion | Chr5 | 20199305 | 20199347 | 42 | |
| Deletion | Chr2 | 22522669 | 22522677 | 8 | |
| Deletion | Chr9 | 22942001 | 23013000 | 71000 | 10 |
| Deletion | Chr3 | 23630340 | 23630341 | 1 | LOC_Os03g42464 |
| Deletion | Chr5 | 24018694 | 24018697 | 3 | |
| Deletion | Chr5 | 25180291 | 25180295 | 4 | LOC_Os05g43280 |
| Deletion | Chr7 | 26410716 | 26410717 | 1 | |
| Deletion | Chr1 | 28581572 | 28581577 | 5 | |
| Deletion | Chr5 | 29074293 | 29074294 | 1 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 2981339 | 2981342 | 4 | LOC_Os10g05900 |
| Insertion | Chr2 | 19305919 | 19305943 | 25 | |
| Insertion | Chr2 | 5398468 | 5398468 | 1 | |
| Insertion | Chr4 | 34970872 | 34970947 | 76 | |
| Insertion | Chr5 | 28034317 | 28034317 | 1 | |
| Insertion | Chr5 | 4452219 | 4452219 | 1 | |
| Insertion | Chr5 | 9621118 | 9621133 | 16 | |
| Insertion | Chr8 | 3535197 | 3535197 | 1 | LOC_Os08g06370 |
| Insertion | Chr9 | 18262261 | 18262261 | 1 | |
| Insertion | Chr9 | 7196896 | 7196897 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 11651642 | 12054298 | LOC_Os03g20550 |
| Inversion | Chr3 | 12028893 | 12054324 | |
| Inversion | Chr8 | 27300756 | 27555057 | LOC_Os08g43560 |
| Inversion | Chr8 | 27300808 | 27555084 | LOC_Os08g43560 |
| Inversion | Chr3 | 32892831 | 33329968 | LOC_Os03g58530 |
No Translocation
