Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1492-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1492-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 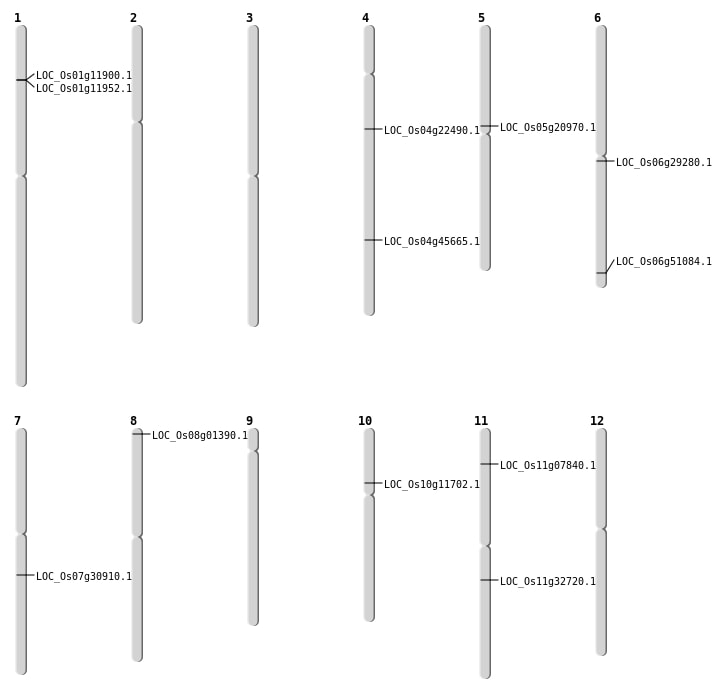
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15793169 | T-G | Homo | INTRON | |
| SBS | Chr1 | 21513536 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 21568547 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 6447656 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g11900 |
| SBS | Chr10 | 1081912 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 19582727 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 6469315 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g11702 |
| SBS | Chr10 | 9391925 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 11408294 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 3997403 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g07840 |
| SBS | Chr11 | 9108404 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 12953484 | T-C | Homo | INTERGENIC | |
| SBS | Chr12 | 8878729 | A-G | Homo | INTERGENIC | |
| SBS | Chr2 | 15018208 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 2806361 | G-A | HET | INTRON | |
| SBS | Chr2 | 6579241 | T-C | HET | INTRON | |
| SBS | Chr3 | 22767835 | C-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr3 | 28501288 | G-A | HET | INTRON | |
| SBS | Chr3 | 29692433 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 32893619 | G-C | Homo | INTERGENIC | |
| SBS | Chr3 | 4161295 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 12733642 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g22490 |
| SBS | Chr4 | 1807749 | C-T | Homo | INTERGENIC | |
| SBS | Chr4 | 18273047 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 27014057 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g45665 |
| SBS | Chr4 | 3932240 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 9704362 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 27545017 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 29407138 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 30901863 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g51084 |
| SBS | Chr6 | 7270723 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 9272466 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 19006643 | G-A | HET | INTRON | |
| SBS | Chr7 | 8108881 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 9643610 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 11433358 | T-G | Homo | INTERGENIC | |
| SBS | Chr8 | 1303493 | C-T | Homo | INTRON | |
| SBS | Chr8 | 17367275 | A-T | Homo | INTERGENIC | |
| SBS | Chr8 | 252852 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g01390 |
| SBS | Chr8 | 27022053 | T-C | Homo | INTRON | |
| SBS | Chr9 | 10455243 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 16885720 | G-A | Homo | INTERGENIC | |
| SBS | Chr9 | 239871 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 3296347 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 4107808 | C-G | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 1207706 | 1207717 | 11 | |
| Deletion | Chr4 | 5149112 | 5149561 | 449 | |
| Deletion | Chr2 | 5675331 | 5675343 | 12 | |
| Deletion | Chr1 | 6513068 | 6513083 | 15 | LOC_Os01g11952 |
| Deletion | Chr1 | 8467896 | 8467925 | 29 | |
| Deletion | Chr5 | 12334220 | 12334226 | 6 | LOC_Os05g20970 |
| Deletion | Chr7 | 15654691 | 15654692 | 1 | |
| Deletion | Chr4 | 16184682 | 16184685 | 3 | |
| Deletion | Chr11 | 16404119 | 16404127 | 8 | |
| Deletion | Chr6 | 16717076 | 16717077 | 1 | LOC_Os06g29280 |
| Deletion | Chr7 | 17415620 | 17415629 | 9 | |
| Deletion | Chr7 | 18300772 | 18300784 | 12 | LOC_Os07g30910 |
| Deletion | Chr11 | 19333197 | 19333213 | 16 | LOC_Os11g32720 |
| Deletion | Chr12 | 19500573 | 19500574 | 1 | |
| Deletion | Chr2 | 21595904 | 21595917 | 13 | |
| Deletion | Chr2 | 25156094 | 25156113 | 19 | |
| Deletion | Chr5 | 25699489 | 25699505 | 16 | |
| Deletion | Chr3 | 27968356 | 27968357 | 1 | |
| Deletion | Chr7 | 28609396 | 28609397 | 1 | |
| Deletion | Chr6 | 29643032 | 29643082 | 50 | |
| Deletion | Chr1 | 30001195 | 30001197 | 2 | |
| Deletion | Chr2 | 35847890 | 35847892 | 2 |
Insertions: 25
No Inversion
No Translocation
