Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1494-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1494-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 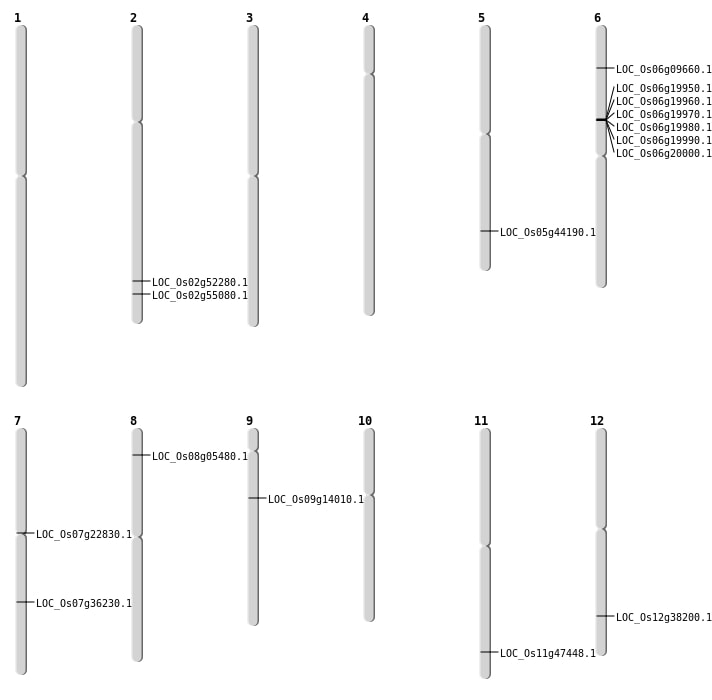
|
| Variant Information |
|---|
Single base substitutions: 25
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15429282 | G-C | HET | INTRON | |
| SBS | Chr1 | 15939713 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 34335478 | C-A | Homo | INTERGENIC | |
| SBS | Chr1 | 34335479 | C-A | Homo | INTERGENIC | |
| SBS | Chr10 | 12994879 | C-T | Homo | INTERGENIC | |
| SBS | Chr11 | 27057130 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 27057131 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 23460323 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g38200 |
| SBS | Chr2 | 32010847 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g52280 |
| SBS | Chr2 | 32641941 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 33676719 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 33735211 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g55080 |
| SBS | Chr2 | 34711752 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 2893819 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 25216932 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 30851457 | G-A | Homo | INTERGENIC | |
| SBS | Chr4 | 5711000 | A-G | Homo | INTERGENIC | |
| SBS | Chr7 | 10033835 | A-G | HET | INTRON | |
| SBS | Chr7 | 14611698 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 14611699 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 602609 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 6460236 | T-C | HET | INTRON | |
| SBS | Chr8 | 12237822 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 15640239 | A-G | HET | INTRON | |
| SBS | Chr8 | 15640529 | A-T | HET | INTRON |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 2920293 | 2920593 | 300 | |
| Deletion | Chr7 | 4145279 | 4145295 | 16 | |
| Deletion | Chr8 | 5045955 | 5045962 | 7 | |
| Deletion | Chr1 | 6288333 | 6288339 | 6 | |
| Deletion | Chr4 | 7613135 | 7613137 | 2 | |
| Deletion | Chr8 | 7779123 | 7779124 | 1 | |
| Deletion | Chr7 | 8773638 | 8773653 | 15 | |
| Deletion | Chr8 | 10167917 | 10167918 | 1 | |
| Deletion | Chr6 | 11659741 | 11659742 | 1 | |
| Deletion | Chr11 | 12045246 | 12045247 | 1 | |
| Deletion | Chr7 | 12876888 | 12878429 | 1541 | LOC_Os07g22830 |
| Deletion | Chr3 | 13517926 | 13517927 | 1 | |
| Deletion | Chr7 | 21656218 | 21656220 | 2 | LOC_Os07g36230 |
| Deletion | Chr6 | 22279433 | 22279438 | 5 | |
| Deletion | Chr3 | 23507367 | 23507381 | 14 | |
| Deletion | Chr6 | 24461320 | 24461405 | 85 | |
| Deletion | Chr5 | 24699586 | 24699587 | 1 | |
| Deletion | Chr5 | 25681932 | 25681937 | 5 | LOC_Os05g44190 |
| Deletion | Chr4 | 27160828 | 27160829 | 1 | |
| Deletion | Chr2 | 27196434 | 27196438 | 4 | |
| Deletion | Chr2 | 27506726 | 27506735 | 9 | |
| Deletion | Chr4 | 31748977 | 31749005 | 28 |
Insertions: 14
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 4308467 | 4927566 | LOC_Os06g09660 |
| Inversion | Chr6 | 4308480 | 4927603 | LOC_Os06g09660 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 8264897 | Chr6 | 10400948 | LOC_Os09g14010 |
| Translocation | Chr11 | 28662509 | Chr8 | 2920306 | 2 |
| Translocation | Chr11 | 28662552 | Chr9 | 8266078 | 2 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr6 | 11424369 | 11469463 | 45094 | 6 |
