Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1496-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1496-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 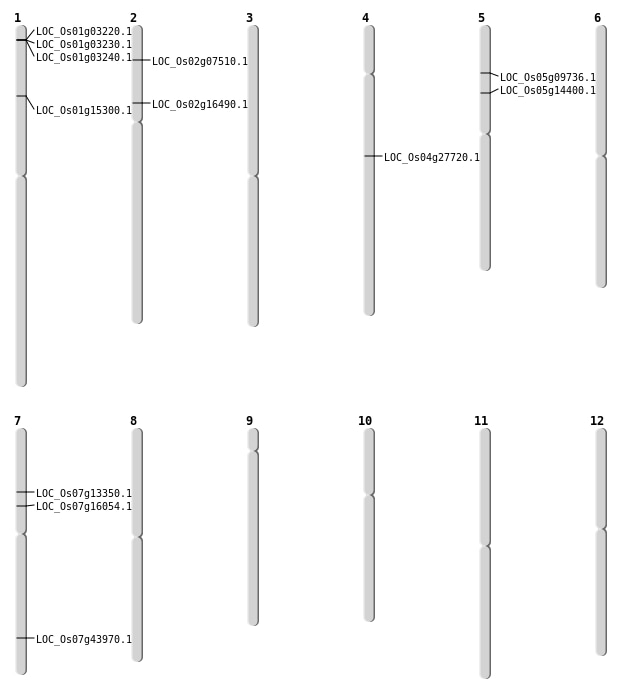
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10802739 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr1 | 13409748 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 8561063 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g15300 |
| SBS | Chr10 | 12056414 | C-A | Homo | SPLICE_SITE_REGION | |
| SBS | Chr10 | 22905633 | G-T | Homo | INTERGENIC | |
| SBS | Chr10 | 5407608 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 23763087 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 2752517 | G-T | Homo | INTERGENIC | |
| SBS | Chr11 | 8089663 | C-T | Homo | INTERGENIC | |
| SBS | Chr2 | 3888033 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g07510 |
| SBS | Chr3 | 1653461 | T-A | Homo | UTR_3_PRIME | |
| SBS | Chr3 | 22566260 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 2535376 | T-C | Homo | INTRON | |
| SBS | Chr3 | 30418321 | G-T | Homo | INTERGENIC | |
| SBS | Chr3 | 9827961 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 16383717 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g27720 |
| SBS | Chr5 | 12597599 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 15387078 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 17393812 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 8112906 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 8112907 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g14400 |
| SBS | Chr6 | 26582877 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 26760017 | G-A | Homo | INTERGENIC | |
| SBS | Chr7 | 29253137 | G-A | Homo | INTERGENIC | |
| SBS | Chr7 | 29253138 | G-A | Homo | INTERGENIC | |
| SBS | Chr7 | 7663724 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g13350 |
| SBS | Chr7 | 7663725 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g13350 |
| SBS | Chr8 | 1518433 | C-A | Homo | INTERGENIC | |
| SBS | Chr8 | 20920669 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 6798092 | C-T | Homo | INTERGENIC | |
| SBS | Chr9 | 21524495 | A-C | HET | INTRON |
Deletions: 38
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1014989 | 1014997 | 8 | |
| Deletion | Chr1 | 1302544 | 1314663 | 12119 | 3 |
| Deletion | Chr3 | 2362475 | 2362476 | 1 | |
| Deletion | Chr3 | 2535154 | 2535164 | 10 | |
| Deletion | Chr5 | 2943967 | 2943976 | 9 | |
| Deletion | Chr10 | 3478370 | 3478371 | 1 | |
| Deletion | Chr5 | 3484472 | 3484473 | 1 | |
| Deletion | Chr10 | 3861913 | 3861914 | 1 | |
| Deletion | Chr5 | 5523632 | 5523642 | 10 | LOC_Os05g09736 |
| Deletion | Chr7 | 5816518 | 5816521 | 3 | |
| Deletion | Chr11 | 5842813 | 5842818 | 5 | |
| Deletion | Chr8 | 6415721 | 6415722 | 1 | |
| Deletion | Chr8 | 6761282 | 6761315 | 33 | |
| Deletion | Chr5 | 7832643 | 7832646 | 3 | |
| Deletion | Chr2 | 9339987 | 9339994 | 7 | |
| Deletion | Chr7 | 9359689 | 9359690 | 1 | LOC_Os07g16054 |
| Deletion | Chr2 | 9378529 | 9378536 | 7 | |
| Deletion | Chr2 | 9410618 | 9410620 | 2 | LOC_Os02g16490 |
| Deletion | Chr3 | 11314137 | 11314139 | 2 | |
| Deletion | Chr4 | 11974382 | 11974384 | 2 | |
| Deletion | Chr4 | 11985288 | 11985290 | 2 | |
| Deletion | Chr10 | 12127607 | 12127613 | 6 | |
| Deletion | Chr11 | 13148274 | 13148276 | 2 | |
| Deletion | Chr7 | 16034596 | 16034597 | 1 | |
| Deletion | Chr9 | 16042369 | 16042370 | 1 | |
| Deletion | Chr1 | 16811714 | 16829303 | 17589 | |
| Deletion | Chr12 | 21465824 | 21465843 | 19 | |
| Deletion | Chr4 | 23152704 | 23152793 | 89 | |
| Deletion | Chr7 | 26289379 | 26289380 | 1 | LOC_Os07g43970 |
| Deletion | Chr8 | 26570461 | 26570462 | 1 | |
| Deletion | Chr2 | 27615669 | 27615676 | 7 | |
| Deletion | Chr11 | 28725109 | 28725117 | 8 | |
| Deletion | Chr5 | 29013408 | 29013412 | 4 | |
| Deletion | Chr3 | 29405916 | 29405919 | 3 | |
| Deletion | Chr1 | 30741810 | 30741812 | 2 | |
| Deletion | Chr3 | 31052754 | 31052756 | 2 | |
| Deletion | Chr1 | 38907239 | 38907241 | 2 | |
| Deletion | Chr1 | 39742288 | 39742290 | 2 |
Insertions: 5
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 14373487 | 17770226 | |
| Inversion | Chr4 | 18861434 | 19144630 |
No Translocation
