Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1497-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1497-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 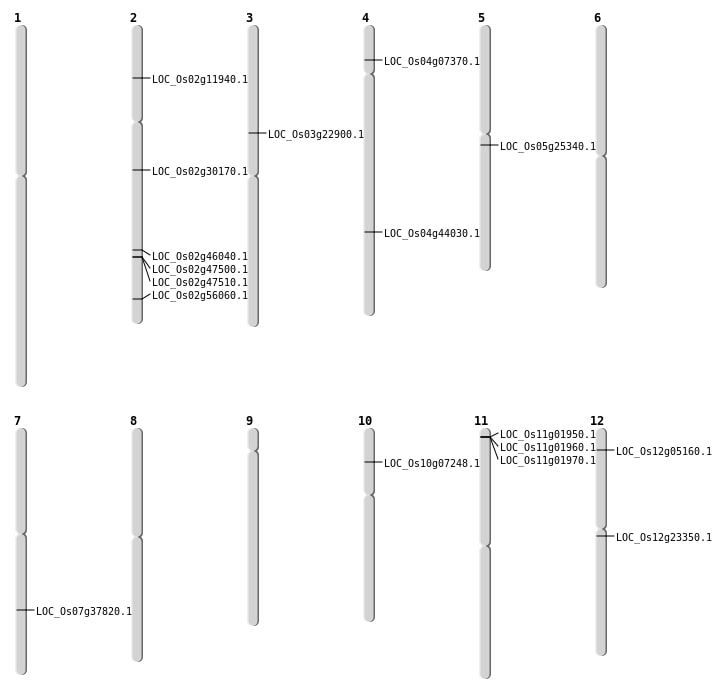
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 31759553 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 3813073 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g07248 |
| SBS | Chr11 | 16936356 | C-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr11 | 17837728 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 22291403 | G-T | Homo | INTRON | |
| SBS | Chr12 | 13217382 | G-T | Homo | STOP_GAINED | LOC_Os12g23350 |
| SBS | Chr12 | 15541007 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 2281230 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g05160 |
| SBS | Chr12 | 2723965 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 25949176 | T-G | HET | INTRON | |
| SBS | Chr2 | 29232510 | A-T | HET | INTRON | |
| SBS | Chr2 | 3959736 | T-A | Homo | INTERGENIC | |
| SBS | Chr2 | 6193169 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g11940 |
| SBS | Chr2 | 6709073 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 25642881 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 29335755 | G-T | HET | UTR_5_PRIME | |
| SBS | Chr3 | 9233155 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 2068377 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 5846098 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 14715132 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g25340 |
| SBS | Chr5 | 29542343 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 4550226 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 5766959 | A-C | HET | UTR_3_PRIME | |
| SBS | Chr7 | 11198254 | G-A | Homo | INTERGENIC | |
| SBS | Chr7 | 22672035 | C-T | HET | SYNONYMOUS_STOP | |
| SBS | Chr7 | 28828542 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 6799760 | T-C | Homo | UTR_3_PRIME | |
| SBS | Chr8 | 27831334 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 8925488 | C-T | HET | INTRON |
Deletions: 41
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 510763 | 522787 | 12024 | 3 |
| Deletion | Chr9 | 3813846 | 3813852 | 6 | |
| Deletion | Chr4 | 3926026 | 3926030 | 4 | LOC_Os04g07370 |
| Deletion | Chr11 | 3941173 | 3941182 | 9 | |
| Deletion | Chr10 | 4967414 | 4967417 | 3 | |
| Deletion | Chr5 | 5947575 | 5947583 | 8 | |
| Deletion | Chr2 | 8153636 | 8153637 | 1 | |
| Deletion | Chr5 | 9256628 | 9256639 | 11 | |
| Deletion | Chr2 | 9699726 | 9699744 | 18 | |
| Deletion | Chr8 | 10016076 | 10016077 | 1 | |
| Deletion | Chr11 | 10228897 | 10228898 | 1 | |
| Deletion | Chr11 | 10565073 | 10565080 | 7 | |
| Deletion | Chr4 | 10896804 | 10896805 | 1 | |
| Deletion | Chr5 | 11728185 | 11728186 | 1 | |
| Deletion | Chr6 | 12772700 | 12772704 | 4 | |
| Deletion | Chr3 | 13242102 | 13242115 | 13 | LOC_Os03g22900 |
| Deletion | Chr6 | 14001593 | 14001683 | 90 | |
| Deletion | Chr12 | 14093830 | 14093852 | 22 | |
| Deletion | Chr11 | 14283594 | 14283595 | 1 | |
| Deletion | Chr1 | 16317247 | 16317305 | 58 | |
| Deletion | Chr1 | 17432666 | 17432667 | 1 | |
| Deletion | Chr1 | 17779523 | 17779524 | 1 | |
| Deletion | Chr2 | 17927524 | 17929167 | 1643 | LOC_Os02g30170 |
| Deletion | Chr2 | 18487730 | 18487742 | 12 | |
| Deletion | Chr12 | 19886785 | 19886798 | 13 | |
| Deletion | Chr4 | 20038052 | 20038054 | 2 | |
| Deletion | Chr11 | 20427021 | 20427026 | 5 | |
| Deletion | Chr6 | 21514970 | 21514971 | 1 | |
| Deletion | Chr7 | 22681256 | 22681260 | 4 | LOC_Os07g37820 |
| Deletion | Chr1 | 23363041 | 23363042 | 1 | |
| Deletion | Chr12 | 23486266 | 23486267 | 1 | |
| Deletion | Chr1 | 24307447 | 24307449 | 2 | |
| Deletion | Chr4 | 26093764 | 26093765 | 1 | LOC_Os04g44030 |
| Deletion | Chr2 | 26804827 | 26804835 | 8 | |
| Deletion | Chr3 | 28305980 | 28305981 | 1 | |
| Deletion | Chr2 | 29024644 | 29027278 | 2634 | 2 |
| Deletion | Chr2 | 29220423 | 29220432 | 9 | |
| Deletion | Chr2 | 29231379 | 29231380 | 1 | |
| Deletion | Chr5 | 29583114 | 29583115 | 1 | |
| Deletion | Chr2 | 34315421 | 34315422 | 1 | LOC_Os02g56060 |
| Deletion | Chr3 | 34705954 | 34705955 | 1 |
Insertions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 20418513 | 20418514 | 2 | |
| Insertion | Chr1 | 42907858 | 42907875 | 18 | |
| Insertion | Chr10 | 2769497 | 2769574 | 78 | |
| Insertion | Chr11 | 22961883 | 22961886 | 4 | |
| Insertion | Chr11 | 7897062 | 7897062 | 1 | |
| Insertion | Chr2 | 28054698 | 28054765 | 68 | LOC_Os02g46040 |
| Insertion | Chr3 | 24454434 | 24454465 | 32 | |
| Insertion | Chr3 | 32638967 | 32639038 | 72 | |
| Insertion | Chr4 | 30424160 | 30424160 | 1 | |
| Insertion | Chr5 | 3125273 | 3125280 | 8 | |
| Insertion | Chr6 | 1477903 | 1477908 | 6 | |
| Insertion | Chr7 | 23454251 | 23454251 | 1 | |
| Insertion | Chr9 | 22493059 | 22493100 | 42 | |
| Insertion | Chr9 | 22800028 | 22800029 | 2 |
No Inversion
No Translocation
