Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1499-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1499-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 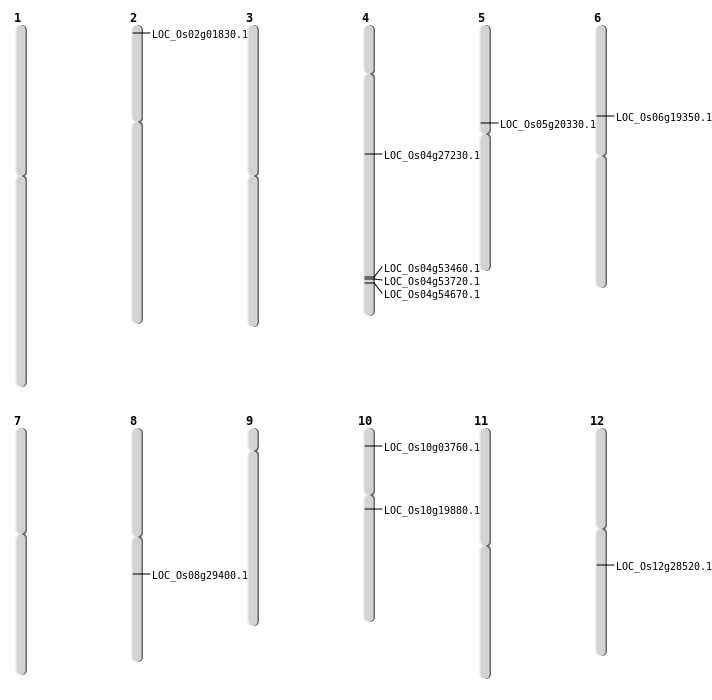
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16918863 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 22555540 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 6896979 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 7748317 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 10952343 | A-T | Homo | INTERGENIC | |
| SBS | Chr10 | 11367829 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 12362232 | G-T | Homo | INTRON | |
| SBS | Chr10 | 1362316 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 4491327 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 7250135 | C-A | Homo | INTERGENIC | |
| SBS | Chr10 | 9921534 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g19880 |
| SBS | Chr11 | 15334701 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 1685295 | T-A | HET | INTRON | |
| SBS | Chr12 | 14747407 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 16346209 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 16649708 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 22557785 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 4549966 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 2308978 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 29878140 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr2 | 31418643 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 32450008 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 33981297 | C-G | Homo | INTERGENIC | |
| SBS | Chr2 | 8427881 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 9281921 | G-A | Homo | INTERGENIC | |
| SBS | Chr3 | 14943237 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 26413680 | A-T | Homo | INTERGENIC | |
| SBS | Chr3 | 34698907 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 1766437 | A-T | Homo | INTERGENIC | |
| SBS | Chr5 | 1974411 | G-A | Homo | UTR_3_PRIME | |
| SBS | Chr5 | 19981432 | A-T | Homo | INTERGENIC | |
| SBS | Chr6 | 11009341 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g19350 |
| SBS | Chr6 | 8074142 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 4004008 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 524749 | A-C | HET | INTRON | |
| SBS | Chr7 | 8719071 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 15130611 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 18300754 | A-T | Homo | INTERGENIC | |
| SBS | Chr9 | 15222221 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 20564720 | A-T | HET | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 145849 | 145878 | 29 | |
| Deletion | Chr2 | 458933 | 458934 | 1 | LOC_Os02g01830 |
| Deletion | Chr1 | 1304854 | 1304856 | 2 | |
| Deletion | Chr9 | 3000010 | 3000016 | 6 | |
| Deletion | Chr1 | 3761729 | 3761731 | 2 | |
| Deletion | Chr2 | 5943281 | 5943286 | 5 | |
| Deletion | Chr4 | 6432217 | 6432234 | 17 | |
| Deletion | Chr7 | 7451715 | 7451717 | 2 | |
| Deletion | Chr1 | 9152909 | 9152910 | 1 | |
| Deletion | Chr11 | 10950969 | 10950970 | 1 | |
| Deletion | Chr7 | 13616580 | 13616581 | 1 | |
| Deletion | Chr5 | 13954113 | 13954114 | 1 | |
| Deletion | Chr12 | 14369120 | 14369140 | 20 | |
| Deletion | Chr2 | 15744913 | 15744914 | 1 | |
| Deletion | Chr2 | 16589453 | 16589454 | 1 | |
| Deletion | Chr8 | 18028098 | 18028108 | 10 | LOC_Os08g29400 |
| Deletion | Chr3 | 18871754 | 18871760 | 6 | |
| Deletion | Chr5 | 20916205 | 20916208 | 3 | |
| Deletion | Chr6 | 21634381 | 21634382 | 1 | |
| Deletion | Chr5 | 22221429 | 22221430 | 1 | |
| Deletion | Chr3 | 22888630 | 22888631 | 1 | |
| Deletion | Chr12 | 25078020 | 25078024 | 4 | |
| Deletion | Chr2 | 25366344 | 25366347 | 3 |
Insertions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 2410478 | 2410553 | 76 | |
| Insertion | Chr1 | 42466279 | 42466316 | 38 | |
| Insertion | Chr10 | 21881493 | 21881494 | 2 | |
| Insertion | Chr11 | 19902316 | 19902317 | 2 | |
| Insertion | Chr11 | 27829812 | 27829825 | 14 | |
| Insertion | Chr12 | 17816234 | 17816278 | 45 | |
| Insertion | Chr12 | 20050528 | 20050528 | 1 | |
| Insertion | Chr12 | 806220 | 806220 | 1 | |
| Insertion | Chr3 | 10751500 | 10751500 | 1 | |
| Insertion | Chr4 | 16094265 | 16094265 | 1 | LOC_Os04g27230 |
| Insertion | Chr4 | 24731227 | 24731227 | 1 | |
| Insertion | Chr5 | 11921199 | 11921233 | 35 | LOC_Os05g20330 |
| Insertion | Chr6 | 3999411 | 3999412 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 1361070 | 1701688 | LOC_Os10g03760 |
| Inversion | Chr10 | 1361086 | 1701702 | LOC_Os10g03760 |
| Inversion | Chr12 | 16395029 | 16852517 | LOC_Os12g28520 |
| Inversion | Chr4 | 31706062 | 32525050 | LOC_Os04g54670 |
| Inversion | Chr4 | 31832092 | 32015748 | 2 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 11421412 | Chr4 | 31830991 | |
| Translocation | Chr10 | 11426729 | Chr4 | 31832033 | LOC_Os04g53460 |
| Translocation | Chr8 | 22080307 | Chr6 | 1069092 |
