Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1505-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1505-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 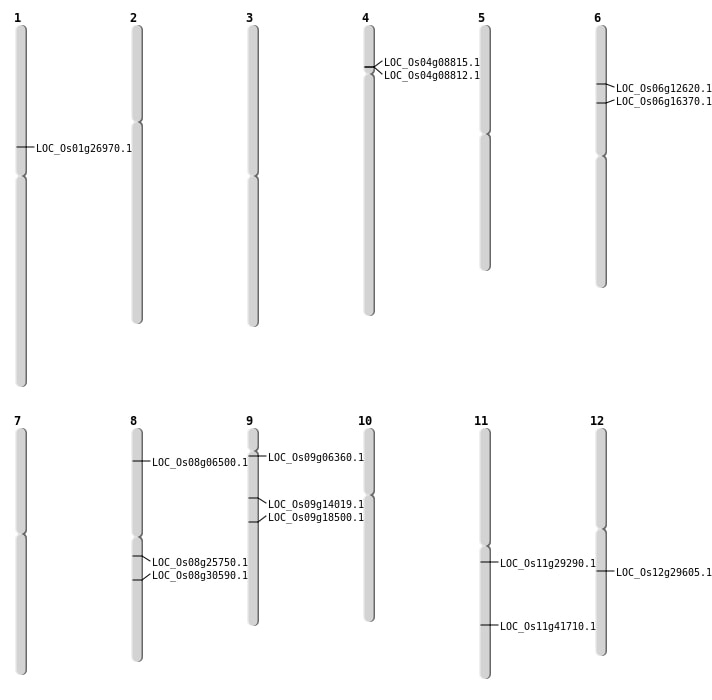
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24609153 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 36293327 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 15668350 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 2724219 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 9534108 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 25042590 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g41710 |
| SBS | Chr11 | 4886097 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 12033303 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 15542227 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 1580816 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 1580818 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 7618299 | C-A | HET | INTRON | |
| SBS | Chr12 | 7641669 | T-C | HET | INTRON | |
| SBS | Chr3 | 13469215 | A-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 5830468 | C-A | HET | INTRON | |
| SBS | Chr3 | 9643239 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 12949922 | T-G | Homo | INTERGENIC | |
| SBS | Chr4 | 20270225 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 12694198 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 13987206 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 5007085 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 13265201 | T-C | HET | SPLICE_SITE_REGION | |
| SBS | Chr6 | 19718481 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 20593752 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 11104999 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 14457635 | T-C | HET | INTRON | |
| SBS | Chr7 | 1939682 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 19465565 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 19465566 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 23386867 | A-G | Homo | INTERGENIC | |
| SBS | Chr7 | 6965784 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 11526654 | C-A | HET | INTRON | |
| SBS | Chr8 | 15679493 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g25750 |
| SBS | Chr8 | 18850788 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g30590 |
| SBS | Chr8 | 19874833 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 11341540 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g18500 |
| SBS | Chr9 | 2992485 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g06360 |
| SBS | Chr9 | 9557576 | T-G | HET | INTERGENIC |
Deletions: 41
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 1991035 | 1991036 | 1 | |
| Deletion | Chr10 | 2203722 | 2203723 | 1 | |
| Deletion | Chr8 | 3688200 | 3688201 | 1 | LOC_Os08g06500 |
| Deletion | Chr8 | 3892830 | 3892832 | 2 | |
| Deletion | Chr12 | 4265695 | 4265697 | 2 | |
| Deletion | Chr10 | 4487752 | 4487756 | 4 | |
| Deletion | Chr6 | 4832097 | 4832098 | 1 | |
| Deletion | Chr4 | 4845538 | 4863698 | 18160 | 2 |
| Deletion | Chr6 | 5824318 | 5824321 | 3 | |
| Deletion | Chr6 | 6638052 | 6638053 | 1 | |
| Deletion | Chr6 | 6878768 | 6878771 | 3 | LOC_Os06g12620 |
| Deletion | Chr11 | 7635710 | 7635734 | 24 | |
| Deletion | Chr1 | 7638507 | 7638508 | 1 | |
| Deletion | Chr12 | 7640410 | 7640413 | 3 | |
| Deletion | Chr7 | 8441495 | 8441496 | 1 | |
| Deletion | Chr6 | 9337034 | 9337052 | 18 | LOC_Os06g16370 |
| Deletion | Chr5 | 10276590 | 10276591 | 1 | |
| Deletion | Chr1 | 11451931 | 11451939 | 8 | |
| Deletion | Chr6 | 14809283 | 14809288 | 5 | |
| Deletion | Chr1 | 15020403 | 15020409 | 6 | LOC_Os01g26970 |
| Deletion | Chr5 | 16117011 | 16117012 | 1 | |
| Deletion | Chr5 | 16117023 | 16117024 | 1 | |
| Deletion | Chr11 | 16986197 | 16986200 | 3 | LOC_Os11g29290 |
| Deletion | Chr8 | 17070277 | 17070298 | 21 | |
| Deletion | Chr12 | 17167271 | 17167272 | 1 | |
| Deletion | Chr6 | 17319843 | 17319845 | 2 | |
| Deletion | Chr6 | 17447287 | 17447306 | 19 | |
| Deletion | Chr1 | 17611686 | 17611688 | 2 | |
| Deletion | Chr12 | 17661072 | 17661073 | 1 | LOC_Os12g29605 |
| Deletion | Chr6 | 18871581 | 18871582 | 1 | |
| Deletion | Chr11 | 19924628 | 19924644 | 16 | |
| Deletion | Chr7 | 20383853 | 20383854 | 1 | |
| Deletion | Chr6 | 20554456 | 20554457 | 1 | |
| Deletion | Chr6 | 21017658 | 21017659 | 1 | |
| Deletion | Chr4 | 22505150 | 22505196 | 46 | |
| Deletion | Chr12 | 25258594 | 25258595 | 1 | |
| Deletion | Chr3 | 25528868 | 25528987 | 119 | |
| Deletion | Chr4 | 25887960 | 25887961 | 1 | |
| Deletion | Chr11 | 26794654 | 26794655 | 1 | |
| Deletion | Chr5 | 29103569 | 29103571 | 2 | |
| Deletion | Chr3 | 29352391 | 29352392 | 1 |
Insertions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 1623145 | 1623146 | 2 | |
| Insertion | Chr11 | 25763332 | 25763335 | 4 | |
| Insertion | Chr12 | 9709792 | 9709813 | 22 | |
| Insertion | Chr4 | 18552929 | 18552929 | 1 | |
| Insertion | Chr4 | 22875321 | 22875321 | 1 | |
| Insertion | Chr4 | 30718755 | 30718756 | 2 | |
| Insertion | Chr5 | 10210194 | 10210194 | 1 | |
| Insertion | Chr5 | 16785370 | 16785373 | 4 | |
| Insertion | Chr5 | 21181961 | 21181962 | 2 | |
| Insertion | Chr6 | 12035929 | 12035929 | 1 | |
| Insertion | Chr6 | 16620877 | 16620878 | 2 | |
| Insertion | Chr7 | 16871129 | 16871129 | 1 | |
| Insertion | Chr7 | 3304066 | 3304081 | 16 | |
| Insertion | Chr8 | 20633622 | 20633623 | 2 | |
| Insertion | Chr9 | 3643799 | 3643837 | 39 | |
| Insertion | Chr9 | 8285447 | 8285447 | 1 | LOC_Os09g14019 |
No Inversion
No Translocation
