Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1508-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1508-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 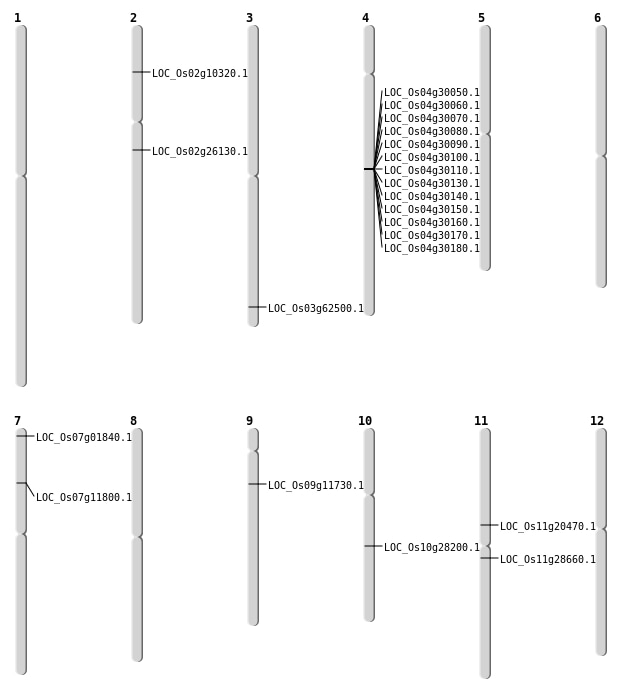
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15263429 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 26896886 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 27277478 | G-T | HET | INTRON | |
| SBS | Chr1 | 34517435 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 14642702 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g28200 |
| SBS | Chr11 | 11856917 | C-T | HET | STOP_GAINED | LOC_Os11g20470 |
| SBS | Chr11 | 18627248 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 21814101 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 25096761 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 13534741 | C-T | Homo | INTRON | |
| SBS | Chr2 | 30370446 | C-T | Homo | INTERGENIC | |
| SBS | Chr2 | 32042403 | A-T | Homo | INTRON | |
| SBS | Chr2 | 548802 | C-A | Homo | INTERGENIC | |
| SBS | Chr3 | 29129302 | G-A | Homo | INTRON | |
| SBS | Chr4 | 12792495 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 33300835 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 34627370 | G-C | HET | INTRON | |
| SBS | Chr4 | 4375989 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 5162542 | A-T | HET | INTRON | |
| SBS | Chr5 | 28359956 | T-C | Homo | INTERGENIC | |
| SBS | Chr5 | 4332934 | G-A | Homo | INTERGENIC | |
| SBS | Chr5 | 6450931 | C-T | Homo | INTRON | |
| SBS | Chr6 | 6341012 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 2435242 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 24739814 | A-G | Homo | INTRON | |
| SBS | Chr7 | 4666351 | T-G | HET | INTRON | |
| SBS | Chr7 | 8404042 | C-T | Homo | INTERGENIC | |
| SBS | Chr8 | 23442305 | G-A | Homo | INTERGENIC | |
| SBS | Chr8 | 7518954 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 14687465 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 6546300 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g11730 |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 498148 | 498149 | 1 | LOC_Os07g01840 |
| Deletion | Chr8 | 3252028 | 3252038 | 10 | |
| Deletion | Chr2 | 5422262 | 5422263 | 1 | LOC_Os02g10320 |
| Deletion | Chr7 | 5670171 | 5670204 | 33 | |
| Deletion | Chr2 | 6010829 | 6010831 | 2 | |
| Deletion | Chr4 | 6016522 | 6016530 | 8 | |
| Deletion | Chr7 | 6531029 | 6531052 | 23 | LOC_Os07g11800 |
| Deletion | Chr11 | 7767725 | 7767726 | 1 | |
| Deletion | Chr9 | 8188321 | 8188325 | 4 | |
| Deletion | Chr3 | 9573359 | 9573360 | 1 | |
| Deletion | Chr7 | 10095333 | 10095334 | 1 | |
| Deletion | Chr11 | 12274595 | 12274596 | 1 | |
| Deletion | Chr8 | 12443654 | 12443657 | 3 | |
| Deletion | Chr7 | 12858589 | 12858591 | 2 | |
| Deletion | Chr1 | 13998737 | 13998738 | 1 | |
| Deletion | Chr9 | 14142754 | 14142755 | 1 | |
| Deletion | Chr4 | 14214158 | 14214165 | 7 | |
| Deletion | Chr9 | 16324308 | 16324310 | 2 | |
| Deletion | Chr11 | 16576392 | 16576400 | 8 | LOC_Os11g28660 |
| Deletion | Chr9 | 16609860 | 16609861 | 1 | |
| Deletion | Chr4 | 17930001 | 18014000 | 84000 | 14 |
| Deletion | Chr5 | 17994383 | 17994390 | 7 | |
| Deletion | Chr4 | 18102292 | 18102293 | 1 | |
| Deletion | Chr2 | 19387791 | 19387801 | 10 | |
| Deletion | Chr4 | 19585731 | 19585736 | 5 | |
| Deletion | Chr7 | 20765514 | 20765516 | 2 | |
| Deletion | Chr7 | 24629876 | 24629886 | 10 | |
| Deletion | Chr12 | 27291929 | 27291930 | 1 | |
| Deletion | Chr3 | 35386029 | 35386036 | 7 | LOC_Os03g62500 |
| Deletion | Chr1 | 43166096 | 43166099 | 3 | LOC_Os01g74530 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 4781732 | 4781737 | 6 | |
| Insertion | Chr2 | 19962420 | 19962420 | 1 | |
| Insertion | Chr7 | 27282369 | 27282370 | 2 | |
| Insertion | Chr7 | 9114721 | 9114721 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 15353557 | 25364044 | LOC_Os02g26130 |
No Translocation
