Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1509-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1509-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 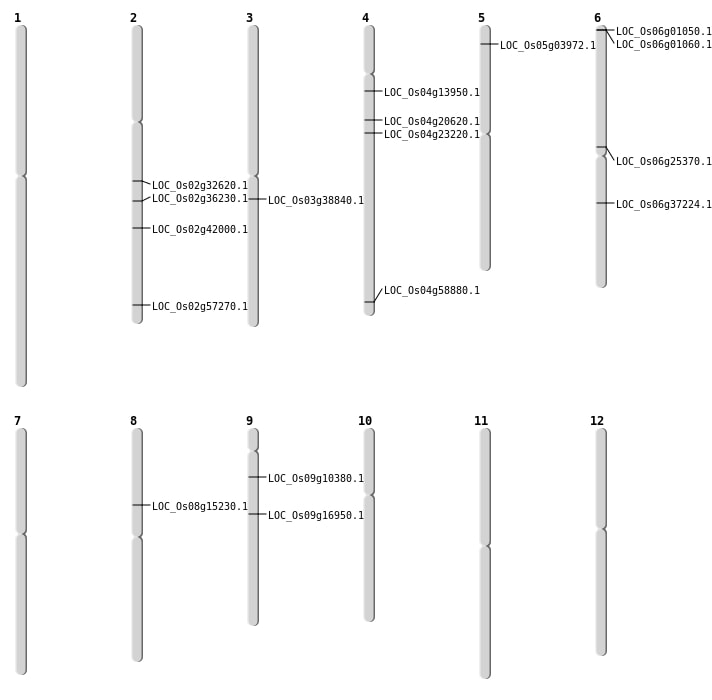
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14949943 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 17726438 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 17726440 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 19502369 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 23082046 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 16914303 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 20976818 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 14576522 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 18944388 | A-C | Homo | INTERGENIC | |
| SBS | Chr11 | 21695794 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 23086598 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 23453604 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 2186916 | A-G | Homo | INTERGENIC | |
| SBS | Chr12 | 25006584 | G-A | HET | INTRON | |
| SBS | Chr2 | 27172870 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 27468024 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 13117761 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 21569645 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g38840 |
| SBS | Chr3 | 28890155 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 35941683 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 15501105 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 22021216 | C-A | Homo | INTRON | |
| SBS | Chr4 | 23601561 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 33067472 | T-C | Homo | INTERGENIC | |
| SBS | Chr4 | 33474184 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 7794476 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g13950 |
| SBS | Chr4 | 8452501 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 12065409 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 14774511 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 21086966 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 12268819 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 14651113 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 14860784 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g25370 |
| SBS | Chr6 | 26446900 | T-A | Homo | INTERGENIC | |
| SBS | Chr7 | 6820605 | T-A | Homo | INTERGENIC | |
| SBS | Chr8 | 17796380 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 18729777 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 5118763 | C-G | HET | UTR_3_PRIME | |
| SBS | Chr8 | 6399989 | C-T | HET | INTRON | |
| SBS | Chr9 | 11600778 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 3194949 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 5681261 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g10380 |
Deletions: 43
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 33975 | 46604 | 12629 | 2 |
| Deletion | Chr7 | 597485 | 597486 | 1 | |
| Deletion | Chr7 | 953948 | 953950 | 2 | |
| Deletion | Chr5 | 1786665 | 1786674 | 9 | LOC_Os05g03972 |
| Deletion | Chr11 | 2891507 | 2891527 | 20 | |
| Deletion | Chr6 | 4456618 | 4456621 | 3 | |
| Deletion | Chr9 | 4555201 | 4555202 | 1 | |
| Deletion | Chr9 | 6345897 | 6345899 | 2 | |
| Deletion | Chr9 | 6793069 | 6793070 | 1 | |
| Deletion | Chr1 | 8307313 | 8307314 | 1 | |
| Deletion | Chr1 | 8538629 | 8538630 | 1 | |
| Deletion | Chr10 | 8735352 | 8735354 | 2 | |
| Deletion | Chr11 | 9027525 | 9027533 | 8 | |
| Deletion | Chr2 | 9031981 | 9031983 | 2 | |
| Deletion | Chr8 | 9224416 | 9224430 | 14 | LOC_Os08g15230 |
| Deletion | Chr4 | 11542059 | 11542062 | 3 | LOC_Os04g20620 |
| Deletion | Chr5 | 11946435 | 11948868 | 2433 | |
| Deletion | Chr6 | 12965264 | 12965265 | 1 | |
| Deletion | Chr4 | 13007643 | 13007644 | 1 | |
| Deletion | Chr4 | 13221356 | 13221371 | 15 | LOC_Os04g23220 |
| Deletion | Chr3 | 13445701 | 13445715 | 14 | |
| Deletion | Chr2 | 13662870 | 13663024 | 154 | |
| Deletion | Chr12 | 14377196 | 14377201 | 5 | |
| Deletion | Chr1 | 16110406 | 16110407 | 1 | |
| Deletion | Chr9 | 16324311 | 16324315 | 4 | |
| Deletion | Chr11 | 17018360 | 17018364 | 4 | |
| Deletion | Chr9 | 18195866 | 18207392 | 11526 | |
| Deletion | Chr5 | 20416168 | 20416172 | 4 | |
| Deletion | Chr2 | 21869040 | 21869041 | 1 | LOC_Os02g36230 |
| Deletion | Chr6 | 21992530 | 21992558 | 28 | LOC_Os06g37224 |
| Deletion | Chr2 | 22522669 | 22522677 | 8 | |
| Deletion | Chr1 | 22745240 | 22745241 | 1 | |
| Deletion | Chr11 | 22749454 | 22749462 | 8 | |
| Deletion | Chr11 | 24348487 | 24348500 | 13 | |
| Deletion | Chr2 | 25232844 | 25232859 | 15 | |
| Deletion | Chr2 | 25239950 | 25239953 | 3 | LOC_Os02g42000 |
| Deletion | Chr8 | 25257558 | 25257559 | 1 | |
| Deletion | Chr5 | 25536204 | 25536234 | 30 | |
| Deletion | Chr12 | 26972901 | 26972903 | 2 | |
| Deletion | Chr7 | 29651492 | 29651493 | 1 | |
| Deletion | Chr4 | 35028057 | 35028060 | 3 | LOC_Os04g58880 |
| Deletion | Chr3 | 35941581 | 35941585 | 4 | |
| Deletion | Chr1 | 38528548 | 38528549 | 1 |
Insertions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 1213455 | 1213463 | 9 | |
| Insertion | Chr1 | 12438573 | 12438573 | 1 | |
| Insertion | Chr10 | 18605247 | 18605306 | 60 | |
| Insertion | Chr10 | 22842543 | 22842543 | 1 | |
| Insertion | Chr11 | 11734176 | 11734178 | 3 | |
| Insertion | Chr12 | 21854734 | 21854747 | 14 | |
| Insertion | Chr12 | 24805688 | 24805688 | 1 | |
| Insertion | Chr2 | 19342067 | 19342072 | 6 | LOC_Os02g32620 |
| Insertion | Chr2 | 35086148 | 35086150 | 3 | LOC_Os02g57270 |
| Insertion | Chr2 | 4921513 | 4921515 | 3 | |
| Insertion | Chr3 | 12114390 | 12114391 | 2 | |
| Insertion | Chr3 | 21407829 | 21407829 | 1 | |
| Insertion | Chr4 | 18330057 | 18330068 | 12 | |
| Insertion | Chr4 | 29034416 | 29034425 | 10 | |
| Insertion | Chr4 | 7256904 | 7256943 | 40 | |
| Insertion | Chr5 | 18214181 | 18214186 | 6 | |
| Insertion | Chr6 | 16134818 | 16134854 | 37 | |
| Insertion | Chr6 | 20528618 | 20528631 | 14 | |
| Insertion | Chr6 | 24037839 | 24037842 | 4 | |
| Insertion | Chr7 | 21545780 | 21545781 | 2 | |
| Insertion | Chr7 | 25221115 | 25221116 | 2 | |
| Insertion | Chr8 | 14823122 | 14823122 | 1 | |
| Insertion | Chr8 | 25824153 | 25824154 | 2 | |
| Insertion | Chr8 | 8657538 | 8657538 | 1 | |
| Insertion | Chr9 | 10365411 | 10365470 | 60 | LOC_Os09g16950 |
| Insertion | Chr9 | 8981681 | 8981682 | 2 |
No Inversion
No Translocation
