Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1515-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1515-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 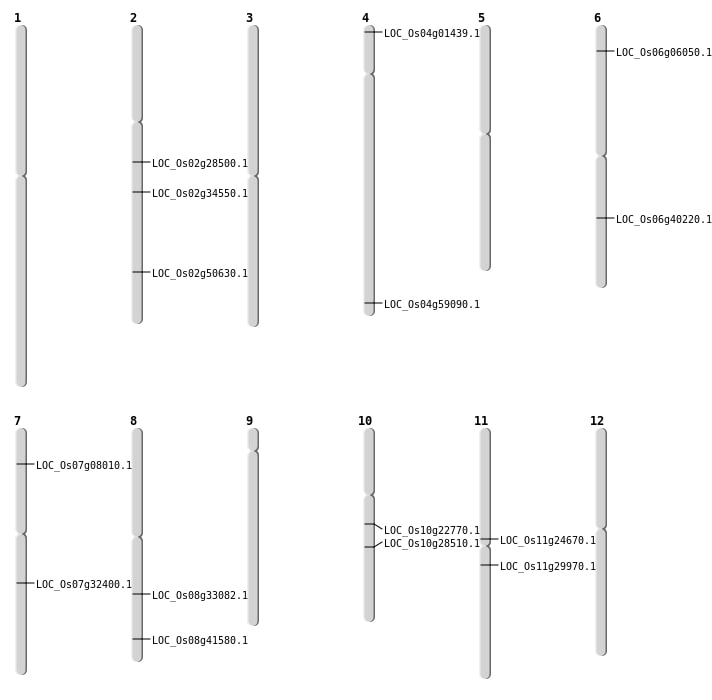
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13850172 | C-A | Homo | INTERGENIC | |
| SBS | Chr1 | 26716254 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 29842797 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr10 | 11837701 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g22770 |
| SBS | Chr11 | 17412532 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g29970 |
| SBS | Chr11 | 18836550 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 19421957 | C-T | Homo | INTERGENIC | |
| SBS | Chr12 | 8337924 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 15246021 | C-T | HET | INTRON | |
| SBS | Chr2 | 2943152 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 30926419 | G-A | HET | STOP_GAINED | LOC_Os02g50630 |
| SBS | Chr2 | 3671083 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 7035881 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 18563763 | A-T | Homo | INTERGENIC | |
| SBS | Chr3 | 6314468 | C-T | Homo | INTERGENIC | |
| SBS | Chr4 | 1390016 | G-A | HET | INTRON | |
| SBS | Chr4 | 17177776 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 33359893 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 5314405 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 13630192 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 24806405 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 23958563 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 23958564 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g40220 |
| SBS | Chr6 | 25261896 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 11198268 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 15177008 | T-C | HET | INTRON | |
| SBS | Chr7 | 15467890 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 26707412 | G-A | HET | INTRON | |
| SBS | Chr7 | 4133317 | T-C | Homo | INTERGENIC | |
| SBS | Chr7 | 6949382 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 10345365 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 14411307 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 21659033 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 13250147 | A-G | Homo | INTERGENIC | |
| SBS | Chr9 | 19957146 | G-T | HET | SYNONYMOUS_CODING |
Deletions: 46
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 298833 | 309180 | 10347 | LOC_Os04g01439 |
| Deletion | Chr10 | 688742 | 688749 | 7 | |
| Deletion | Chr4 | 1903613 | 1903615 | 2 | |
| Deletion | Chr8 | 2355558 | 2355562 | 4 | |
| Deletion | Chr12 | 2716357 | 2716363 | 6 | |
| Deletion | Chr7 | 4058912 | 4058927 | 15 | LOC_Os07g08010 |
| Deletion | Chr2 | 4258004 | 4258008 | 4 | |
| Deletion | Chr7 | 5399526 | 5399529 | 3 | |
| Deletion | Chr4 | 5546389 | 5546390 | 1 | |
| Deletion | Chr9 | 5565609 | 5565612 | 3 | |
| Deletion | Chr2 | 6030286 | 6030287 | 1 | |
| Deletion | Chr1 | 6517777 | 6517803 | 26 | |
| Deletion | Chr8 | 6519399 | 6519401 | 2 | |
| Deletion | Chr4 | 6612751 | 6612765 | 14 | |
| Deletion | Chr11 | 7286199 | 7286200 | 1 | |
| Deletion | Chr7 | 8945749 | 8945757 | 8 | |
| Deletion | Chr7 | 13230596 | 13230620 | 24 | |
| Deletion | Chr11 | 13627460 | 13627461 | 1 | |
| Deletion | Chr9 | 13652327 | 13652335 | 8 | |
| Deletion | Chr9 | 13802956 | 13802957 | 1 | |
| Deletion | Chr6 | 14029804 | 14029806 | 2 | |
| Deletion | Chr11 | 14070776 | 14070777 | 1 | LOC_Os11g24670 |
| Deletion | Chr10 | 14833490 | 14833491 | 1 | LOC_Os10g28510 |
| Deletion | Chr3 | 15610475 | 15610477 | 2 | |
| Deletion | Chr1 | 16857215 | 16857216 | 1 | |
| Deletion | Chr3 | 17323940 | 17323961 | 21 | |
| Deletion | Chr2 | 18320145 | 18320146 | 1 | |
| Deletion | Chr6 | 18695660 | 18695661 | 1 | |
| Deletion | Chr7 | 19262667 | 19262668 | 1 | LOC_Os07g32400 |
| Deletion | Chr11 | 20228227 | 20228233 | 6 | |
| Deletion | Chr8 | 20564578 | 20564580 | 2 | LOC_Os08g33082 |
| Deletion | Chr2 | 20710918 | 20710934 | 16 | LOC_Os02g34550 |
| Deletion | Chr1 | 22233453 | 22233458 | 5 | |
| Deletion | Chr8 | 22544442 | 22544443 | 1 | |
| Deletion | Chr10 | 22611149 | 22611159 | 10 | |
| Deletion | Chr6 | 23372705 | 23372706 | 1 | |
| Deletion | Chr1 | 23825350 | 23825390 | 40 | |
| Deletion | Chr2 | 24022495 | 24022503 | 8 | |
| Deletion | Chr1 | 24408279 | 24408307 | 28 | |
| Deletion | Chr6 | 24865646 | 24865662 | 16 | |
| Deletion | Chr8 | 25192710 | 25192711 | 1 | |
| Deletion | Chr8 | 26266329 | 26266331 | 2 | LOC_Os08g41580 |
| Deletion | Chr4 | 26593257 | 26593259 | 2 | |
| Deletion | Chr1 | 26821219 | 26821295 | 76 | |
| Deletion | Chr3 | 34366426 | 34366429 | 3 | |
| Deletion | Chr4 | 35146137 | 35146138 | 1 | LOC_Os04g59090 |
Insertions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11509713 | 11509715 | 3 | |
| Insertion | Chr11 | 12088883 | 12088935 | 53 | |
| Insertion | Chr11 | 1878359 | 1878360 | 2 | |
| Insertion | Chr11 | 22634284 | 22634285 | 2 | |
| Insertion | Chr11 | 28782639 | 28782640 | 2 | |
| Insertion | Chr11 | 5773089 | 5773111 | 23 | |
| Insertion | Chr2 | 16857280 | 16857297 | 18 | LOC_Os02g28500 |
| Insertion | Chr4 | 20122606 | 20122614 | 9 | |
| Insertion | Chr5 | 21533926 | 21533926 | 1 | |
| Insertion | Chr5 | 49552 | 49553 | 2 | |
| Insertion | Chr6 | 18507252 | 18507253 | 2 | |
| Insertion | Chr6 | 2780756 | 2780757 | 2 | LOC_Os06g06050 |
| Insertion | Chr6 | 3459409 | 3459409 | 1 | |
| Insertion | Chr6 | 899441 | 899441 | 1 | |
| Insertion | Chr7 | 14259631 | 14259689 | 59 | |
| Insertion | Chr7 | 654349 | 654362 | 14 | |
| Insertion | Chr7 | 9430294 | 9430295 | 2 |
No Inversion
No Translocation
