Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1516-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1516-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 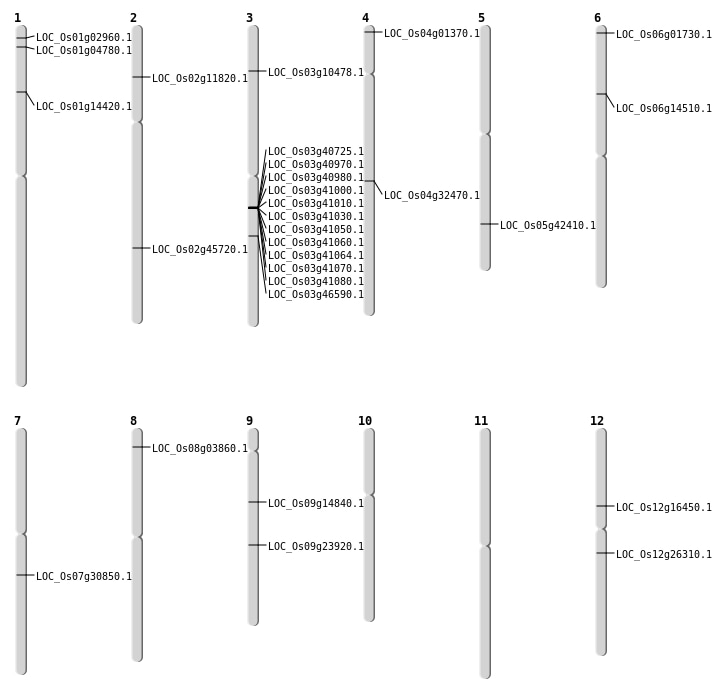
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 23925481 | G-A | Homo | INTRON | |
| SBS | Chr1 | 40015583 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 40238067 | G-A | Homo | UTR_3_PRIME | |
| SBS | Chr1 | 8067011 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g14420 |
| SBS | Chr10 | 14604688 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 11468824 | T-G | Homo | UTR_3_PRIME | |
| SBS | Chr12 | 15356719 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g26310 |
| SBS | Chr12 | 25599175 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 25599176 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 4115463 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 5788951 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 10738237 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 10902876 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 22163189 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 14119638 | C-A | Homo | INTRON | |
| SBS | Chr3 | 15609684 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 3721770 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 5068311 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 5431428 | G-A | HET | INTRON | |
| SBS | Chr4 | 15242428 | T-G | Homo | INTRON | |
| SBS | Chr4 | 26353955 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 27544994 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr4 | 277372 | C-T | HET | STOP_GAINED | LOC_Os04g01370 |
| SBS | Chr6 | 19628509 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 6968587 | G-A | Homo | INTERGENIC | |
| SBS | Chr7 | 10791173 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 18260666 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30850 |
| SBS | Chr7 | 28510712 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 10116074 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 19668533 | T-G | HET | INTRON | |
| SBS | Chr8 | 22068428 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 23667308 | A-T | Homo | INTERGENIC | |
| SBS | Chr8 | 25797826 | T-G | HET | INTERGENIC |
Deletions: 39
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 427663 | 427664 | 1 | LOC_Os06g01730 |
| Deletion | Chr1 | 2187920 | 2187921 | 1 | LOC_Os01g04780 |
| Deletion | Chr5 | 4281740 | 4281745 | 5 | |
| Deletion | Chr3 | 5331423 | 5331427 | 4 | LOC_Os03g10478 |
| Deletion | Chr2 | 6114433 | 6114439 | 6 | LOC_Os02g11820 |
| Deletion | Chr7 | 6976148 | 6976170 | 22 | |
| Deletion | Chr9 | 7043915 | 7043916 | 1 | |
| Deletion | Chr1 | 8064233 | 8064235 | 2 | |
| Deletion | Chr2 | 8481912 | 8481921 | 9 | |
| Deletion | Chr9 | 8846809 | 8846884 | 75 | LOC_Os09g14840 |
| Deletion | Chr5 | 8955269 | 8955274 | 5 | |
| Deletion | Chr4 | 9270412 | 9270432 | 20 | |
| Deletion | Chr12 | 9423591 | 9423599 | 8 | LOC_Os12g16450 |
| Deletion | Chr11 | 11264977 | 11264987 | 10 | |
| Deletion | Chr5 | 11475435 | 11475438 | 3 | |
| Deletion | Chr3 | 12222693 | 12222694 | 1 | |
| Deletion | Chr6 | 12949651 | 12949652 | 1 | |
| Deletion | Chr11 | 13862778 | 13862779 | 1 | |
| Deletion | Chr10 | 14758535 | 14758536 | 1 | |
| Deletion | Chr11 | 15504954 | 15504955 | 1 | |
| Deletion | Chr9 | 16116159 | 16116161 | 2 | |
| Deletion | Chr4 | 18650222 | 18650226 | 4 | |
| Deletion | Chr4 | 19484078 | 19484105 | 27 | LOC_Os04g32470 |
| Deletion | Chr12 | 20367485 | 20367486 | 1 | |
| Deletion | Chr1 | 20584427 | 20584429 | 2 | |
| Deletion | Chr6 | 20609623 | 20609632 | 9 | |
| Deletion | Chr3 | 21047348 | 21047349 | 1 | |
| Deletion | Chr3 | 22770001 | 22842000 | 72000 | 10 |
| Deletion | Chr3 | 23427662 | 23427663 | 1 | |
| Deletion | Chr6 | 24915674 | 24915675 | 1 | |
| Deletion | Chr4 | 25113079 | 25113080 | 1 | |
| Deletion | Chr6 | 26102884 | 26102887 | 3 | |
| Deletion | Chr3 | 26372540 | 26372541 | 1 | LOC_Os03g46590 |
| Deletion | Chr4 | 26725853 | 26725854 | 1 | |
| Deletion | Chr2 | 26991086 | 26991107 | 21 | |
| Deletion | Chr2 | 27818252 | 27818253 | 1 | LOC_Os02g45720 |
| Deletion | Chr7 | 27994281 | 27994282 | 1 | |
| Deletion | Chr1 | 33133950 | 33133954 | 4 | |
| Deletion | Chr1 | 37280819 | 37280825 | 6 |
Insertions: 9
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 1106556 | 1106556 | 1 | LOC_Os01g02960 |
| Insertion | Chr10 | 11296815 | 11296818 | 4 | |
| Insertion | Chr10 | 7034983 | 7034988 | 6 | |
| Insertion | Chr11 | 12297659 | 12297696 | 38 | |
| Insertion | Chr3 | 24154913 | 24154915 | 3 | |
| Insertion | Chr4 | 20111015 | 20111085 | 71 | |
| Insertion | Chr8 | 1840471 | 1840541 | 71 | LOC_Os08g03860 |
| Insertion | Chr9 | 14265569 | 14265571 | 3 | LOC_Os09g23920 |
| Insertion | Chr9 | 17265060 | 17265060 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 8155492 | 8227878 | LOC_Os06g14510 |
| Inversion | Chr3 | 22661045 | 22842348 | LOC_Os03g40725 |
| Inversion | Chr5 | 24840063 | 24896788 | LOC_Os05g42410 |
No Translocation
