Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1519-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1519-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 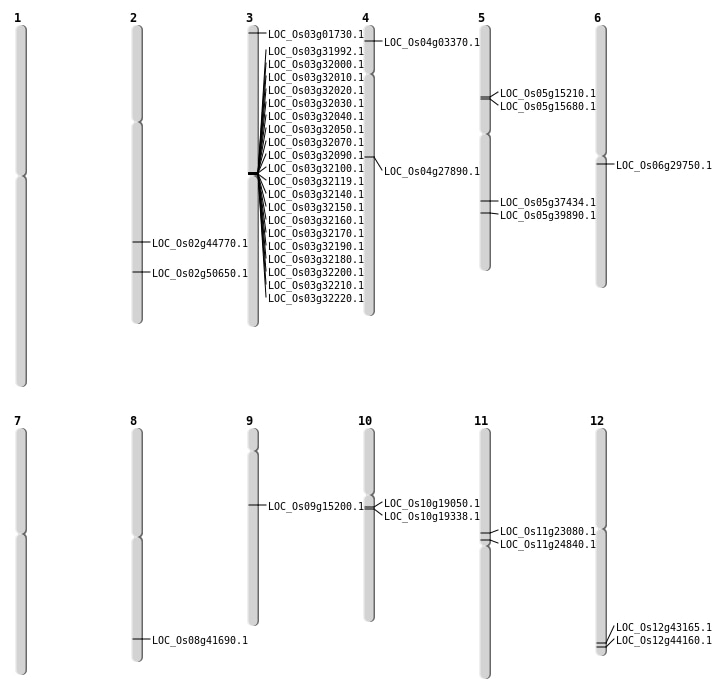
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13318692 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 31681385 | A-T | Homo | UTR_3_PRIME | |
| SBS | Chr10 | 9700328 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g19050 |
| SBS | Chr10 | 9872516 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g19338 |
| SBS | Chr11 | 14164428 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g24840 |
| SBS | Chr11 | 27660284 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 6106691 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 10917642 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 21207814 | A-G | HET | INTRON | |
| SBS | Chr12 | 26806715 | A-G | Homo | START_GAINED | LOC_Os12g43165 |
| SBS | Chr12 | 4732579 | C-T | HET | INTRON | |
| SBS | Chr12 | 6791965 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 27118368 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g44770 |
| SBS | Chr2 | 27118369 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 30941934 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g50650 |
| SBS | Chr2 | 30941955 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g50650 |
| SBS | Chr2 | 6959844 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr3 | 4379581 | A-T | Homo | INTERGENIC | |
| SBS | Chr4 | 10143065 | G-A | Homo | INTERGENIC | |
| SBS | Chr4 | 17414566 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 2125664 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 22642831 | C-T | Homo | INTERGENIC | |
| SBS | Chr4 | 23294886 | T-A | HET | INTRON | |
| SBS | Chr4 | 2386012 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 8969313 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 16367154 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 21532963 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 21905254 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g37434 |
| SBS | Chr5 | 23858587 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 3009658 | A-T | HET | INTRON | |
| SBS | Chr5 | 8785786 | A-G | HET | INTRON | |
| SBS | Chr6 | 17306818 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 19127480 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 4756704 | T-C | Homo | INTRON | |
| SBS | Chr8 | 10337039 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 27501621 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 7598552 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 8261775 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 9599487 | C-G | HET | INTERGENIC |
Deletions: 38
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1447090 | 1447096 | 6 | LOC_Os04g03370 |
| Deletion | Chr1 | 3193320 | 3193321 | 1 | |
| Deletion | Chr11 | 4620325 | 4620326 | 1 | |
| Deletion | Chr11 | 6107858 | 6107859 | 1 | |
| Deletion | Chr5 | 8869962 | 8869967 | 5 | LOC_Os05g15680 |
| Deletion | Chr1 | 9191516 | 9191522 | 6 | |
| Deletion | Chr10 | 9753750 | 9753757 | 7 | |
| Deletion | Chr9 | 10476942 | 10476945 | 3 | |
| Deletion | Chr5 | 11841265 | 11841266 | 1 | |
| Deletion | Chr7 | 13171706 | 13171707 | 1 | |
| Deletion | Chr12 | 13236870 | 13236884 | 14 | |
| Deletion | Chr10 | 13563861 | 13563862 | 1 | |
| Deletion | Chr1 | 13685502 | 13685503 | 1 | |
| Deletion | Chr6 | 14735468 | 14735479 | 11 | |
| Deletion | Chr10 | 15584438 | 15584444 | 6 | |
| Deletion | Chr9 | 16062107 | 16062114 | 7 | |
| Deletion | Chr10 | 16260224 | 16260226 | 2 | |
| Deletion | Chr10 | 16535484 | 16535500 | 16 | |
| Deletion | Chr6 | 17085439 | 17085447 | 8 | LOC_Os06g29750 |
| Deletion | Chr10 | 17406342 | 17406354 | 12 | |
| Deletion | Chr3 | 17769952 | 17769957 | 5 | |
| Deletion | Chr3 | 18300001 | 18435000 | 135000 | 20 |
| Deletion | Chr1 | 18969494 | 18969513 | 19 | |
| Deletion | Chr8 | 20031698 | 20031703 | 5 | |
| Deletion | Chr5 | 21954505 | 21954522 | 17 | |
| Deletion | Chr5 | 23438098 | 23438100 | 2 | LOC_Os05g39890 |
| Deletion | Chr1 | 25985294 | 25985296 | 2 | |
| Deletion | Chr8 | 26315311 | 26315313 | 2 | LOC_Os08g41690 |
| Deletion | Chr12 | 27162961 | 27163028 | 67 | |
| Deletion | Chr6 | 27347764 | 27347765 | 1 | |
| Deletion | Chr8 | 27409831 | 27409842 | 11 | |
| Deletion | Chr4 | 27744669 | 27744677 | 8 | |
| Deletion | Chr4 | 28386697 | 28386700 | 3 | |
| Deletion | Chr7 | 28916743 | 28916744 | 1 | |
| Deletion | Chr7 | 28975265 | 28975266 | 1 | |
| Deletion | Chr3 | 29352144 | 29352148 | 4 | |
| Deletion | Chr3 | 36105554 | 36105566 | 12 | |
| Deletion | Chr1 | 37825004 | 37825010 | 6 |
Insertions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 18619721 | 18619770 | 50 | |
| Insertion | Chr10 | 15018748 | 15018763 | 16 | |
| Insertion | Chr10 | 2675649 | 2675680 | 32 | |
| Insertion | Chr10 | 2778969 | 2778976 | 8 | |
| Insertion | Chr11 | 7559858 | 7559863 | 6 | |
| Insertion | Chr2 | 15667515 | 15667530 | 16 | |
| Insertion | Chr2 | 6394878 | 6394879 | 2 | |
| Insertion | Chr3 | 24268347 | 24268354 | 8 | |
| Insertion | Chr3 | 26960155 | 26960156 | 2 | |
| Insertion | Chr3 | 466208 | 466209 | 2 | LOC_Os03g01730 |
| Insertion | Chr3 | 5965993 | 5966008 | 16 | |
| Insertion | Chr4 | 16473545 | 16473549 | 5 | LOC_Os04g27890 |
| Insertion | Chr4 | 32623545 | 32623550 | 6 | |
| Insertion | Chr6 | 2271224 | 2271235 | 12 | |
| Insertion | Chr8 | 21911113 | 21911114 | 2 | |
| Insertion | Chr9 | 7088190 | 7088197 | 8 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 8632354 | 9089195 | LOC_Os05g15210 |
| Inversion | Chr9 | 8670909 | 9230845 | LOC_Os09g15200 |
| Inversion | Chr9 | 8675173 | 9230846 | LOC_Os09g15200 |
| Inversion | Chr11 | 13281074 | 13285550 | LOC_Os11g23080 |
| Inversion | Chr12 | 27394412 | 27445041 | LOC_Os12g44160 |
| Inversion | Chr12 | 27394423 | 27445054 | LOC_Os12g44160 |
No Translocation
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr8 | 12129869 | 12129968 | 99 |
