Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1521-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1521-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 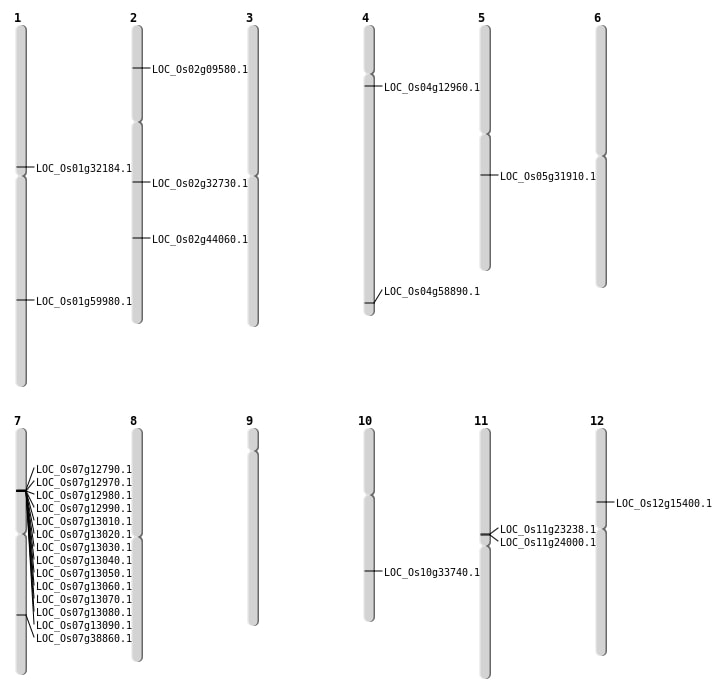
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17620432 | A-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g32184 |
| SBS | Chr1 | 34678425 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g59980 |
| SBS | Chr1 | 34966303 | T-A | Homo | INTERGENIC | |
| SBS | Chr1 | 34966312 | G-C | Homo | INTERGENIC | |
| SBS | Chr1 | 8358521 | C-G | Homo | INTERGENIC | |
| SBS | Chr10 | 2480570 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr10 | 4856174 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 10704964 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 21732777 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr11 | 9270327 | G-A | HET | INTRON | |
| SBS | Chr12 | 16737844 | G-T | Homo | INTERGENIC | |
| SBS | Chr12 | 17805113 | G-C | Homo | INTERGENIC | |
| SBS | Chr2 | 21978201 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 32641964 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 18672999 | A-T | Homo | INTERGENIC | |
| SBS | Chr3 | 3344586 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 9013114 | C-A | Homo | INTRON | |
| SBS | Chr4 | 10934932 | G-C | HET | INTRON | |
| SBS | Chr4 | 10983798 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 2085039 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 26797092 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 4524730 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 7155682 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g12960 |
| SBS | Chr5 | 16079678 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 21590194 | G-A | HET | INTRON | |
| SBS | Chr5 | 21951982 | A-T | HET | INTRON | |
| SBS | Chr6 | 26062501 | G-A | HET | INTRON | |
| SBS | Chr6 | 4838299 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 13898192 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 14281446 | C-A | HET | INTRON | |
| SBS | Chr7 | 23314818 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g38860 |
| SBS | Chr7 | 23314844 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 27319728 | A-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 9592002 | G-T | HET | INTRON | |
| SBS | Chr8 | 25205434 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 5161938 | A-T | HET | INTERGENIC |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 548639 | 548658 | 19 | |
| Deletion | Chr8 | 2225406 | 2225409 | 3 | |
| Deletion | Chr7 | 3518933 | 3518940 | 7 | |
| Deletion | Chr2 | 3588139 | 3588169 | 30 | |
| Deletion | Chr11 | 4158959 | 4158961 | 2 | |
| Deletion | Chr1 | 4831278 | 4831282 | 4 | |
| Deletion | Chr2 | 4933473 | 4933483 | 10 | LOC_Os02g09580 |
| Deletion | Chr11 | 6503181 | 6503187 | 6 | |
| Deletion | Chr7 | 7437001 | 7499000 | 62000 | 11 |
| Deletion | Chr12 | 8797140 | 8797142 | 2 | LOC_Os12g15400 |
| Deletion | Chr7 | 9433152 | 9433153 | 1 | |
| Deletion | Chr10 | 14490066 | 14490072 | 6 | |
| Deletion | Chr9 | 16467660 | 16467662 | 2 | |
| Deletion | Chr7 | 17073307 | 17073326 | 19 | |
| Deletion | Chr5 | 18577061 | 18577223 | 162 | LOC_Os05g31910 |
| Deletion | Chr5 | 19333679 | 19333680 | 1 | |
| Deletion | Chr2 | 19437747 | 19437748 | 1 | LOC_Os02g32730 |
| Deletion | Chr5 | 19917282 | 19917295 | 13 | |
| Deletion | Chr10 | 23182674 | 23182675 | 1 | |
| Deletion | Chr5 | 24649166 | 24649168 | 2 | |
| Deletion | Chr3 | 25955259 | 25955260 | 1 | |
| Deletion | Chr5 | 26296550 | 26296553 | 3 | |
| Deletion | Chr2 | 26593153 | 26593155 | 2 | LOC_Os02g44060 |
| Deletion | Chr7 | 27614441 | 27614446 | 5 | |
| Deletion | Chr1 | 29472937 | 29472938 | 1 | |
| Deletion | Chr4 | 29688265 | 29688269 | 4 | |
| Deletion | Chr1 | 29873318 | 29873320 | 2 | |
| Deletion | Chr1 | 33096669 | 33096670 | 1 | |
| Deletion | Chr4 | 35032052 | 35032092 | 40 | LOC_Os04g58890 |
| Deletion | Chr2 | 35863415 | 35863416 | 1 | |
| Deletion | Chr1 | 42973030 | 42973053 | 23 |
Insertions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11712423 | 11712423 | 1 | |
| Insertion | Chr1 | 21726610 | 21726612 | 3 | |
| Insertion | Chr1 | 9445153 | 9445154 | 2 | |
| Insertion | Chr10 | 17822556 | 17822556 | 1 | LOC_Os10g33740 |
| Insertion | Chr11 | 13435189 | 13435230 | 42 | |
| Insertion | Chr2 | 9452252 | 9452252 | 1 | |
| Insertion | Chr4 | 6510891 | 6510893 | 3 | |
| Insertion | Chr4 | 6690799 | 6690800 | 2 | |
| Insertion | Chr4 | 7765631 | 7765631 | 1 | |
| Insertion | Chr6 | 26612099 | 26612106 | 8 | |
| Insertion | Chr8 | 25412077 | 25412078 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 7317977 | 7437665 | LOC_Os07g12790 |
| Inversion | Chr7 | 7318005 | 7499343 | 2 |
| Inversion | Chr11 | 13402305 | 13600433 | 2 |
| Inversion | Chr8 | 16899686 | 20420969 |
No Translocation
