Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1525-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1525-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 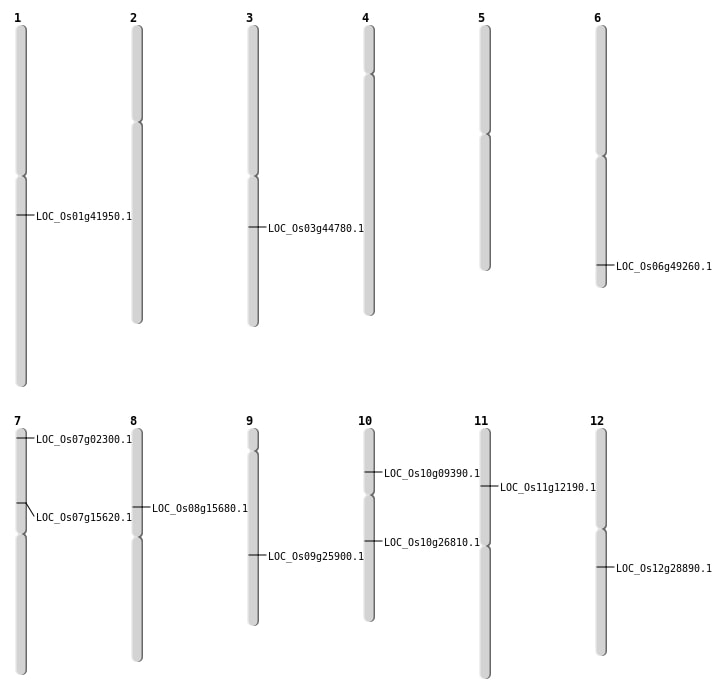
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 32529089 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 6688311 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 112595 | C-T | HET | INTRON | |
| SBS | Chr10 | 15824395 | T-A | Homo | INTERGENIC | |
| SBS | Chr10 | 18336562 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 2303604 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 8025425 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 18104603 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 21400846 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 8965395 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 17082217 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g28890 |
| SBS | Chr12 | 18062096 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 21942756 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 15097934 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 1799984 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 33850366 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 21209588 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 22149135 | C-T | HET | INTRON | |
| SBS | Chr3 | 31102186 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 327678 | A-G | Homo | INTERGENIC | |
| SBS | Chr4 | 13172648 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 23710644 | G-A | Homo | INTERGENIC | |
| SBS | Chr4 | 28550098 | A-T | Homo | INTERGENIC | |
| SBS | Chr4 | 6427384 | G-A | HET | INTRON | |
| SBS | Chr5 | 15491106 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 17100892 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 7008943 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 3719328 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 2259251 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 755823 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g02300 |
| SBS | Chr7 | 755824 | C-A | HET | STOP_GAINED | LOC_Os07g02300 |
| SBS | Chr7 | 861909 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 11836743 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 15009628 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr8 | 15221859 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 21499304 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 13415710 | C-T | HET | INTERGENIC |
Deletions: 39
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 788524 | 788534 | 10 | |
| Deletion | Chr4 | 1683502 | 1683504 | 2 | |
| Deletion | Chr6 | 2756122 | 2756124 | 2 | |
| Deletion | Chr10 | 4360423 | 4360433 | 10 | |
| Deletion | Chr12 | 4670650 | 4670651 | 1 | |
| Deletion | Chr11 | 6794808 | 6794811 | 3 | LOC_Os11g12190 |
| Deletion | Chr11 | 7773279 | 7773281 | 2 | |
| Deletion | Chr7 | 9058642 | 9058643 | 1 | LOC_Os07g15620 |
| Deletion | Chr8 | 9522621 | 9522622 | 1 | LOC_Os08g15680 |
| Deletion | Chr10 | 14047336 | 14047338 | 2 | LOC_Os10g26810 |
| Deletion | Chr8 | 14370800 | 14370804 | 4 | |
| Deletion | Chr9 | 15546550 | 15546557 | 7 | LOC_Os09g25900 |
| Deletion | Chr1 | 16317247 | 16317305 | 58 | |
| Deletion | Chr7 | 17194166 | 17194174 | 8 | |
| Deletion | Chr10 | 17624523 | 17624541 | 18 | |
| Deletion | Chr11 | 17624593 | 17624594 | 1 | |
| Deletion | Chr12 | 17822942 | 17822943 | 1 | |
| Deletion | Chr7 | 17906283 | 17906285 | 2 | |
| Deletion | Chr3 | 20115929 | 20115931 | 2 | |
| Deletion | Chr12 | 20382680 | 20382681 | 1 | |
| Deletion | Chr11 | 20401117 | 20401123 | 6 | |
| Deletion | Chr1 | 20447044 | 20447046 | 2 | |
| Deletion | Chr11 | 21756595 | 21756600 | 5 | |
| Deletion | Chr7 | 21813145 | 21813146 | 1 | |
| Deletion | Chr12 | 22113956 | 22113962 | 6 | |
| Deletion | Chr10 | 23051549 | 23051578 | 29 | |
| Deletion | Chr2 | 23365183 | 23365198 | 15 | |
| Deletion | Chr3 | 25253494 | 25253505 | 11 | LOC_Os03g44780 |
| Deletion | Chr6 | 25993801 | 25993802 | 1 | |
| Deletion | Chr3 | 26125829 | 26125839 | 10 | |
| Deletion | Chr5 | 27370286 | 27370288 | 2 | |
| Deletion | Chr1 | 27726933 | 27726939 | 6 | |
| Deletion | Chr2 | 28497057 | 28497070 | 13 | |
| Deletion | Chr1 | 28507554 | 28507555 | 1 | |
| Deletion | Chr6 | 29226533 | 29226536 | 3 | |
| Deletion | Chr6 | 29854997 | 29855008 | 11 | LOC_Os06g49260 |
| Deletion | Chr4 | 32486210 | 32486223 | 13 | |
| Deletion | Chr2 | 34837057 | 34837069 | 12 | |
| Deletion | Chr2 | 35130339 | 35130340 | 1 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 10065547 | 10065576 | 30 | |
| Insertion | Chr1 | 23783017 | 23783062 | 46 | LOC_Os01g41950 |
| Insertion | Chr1 | 4006192 | 4006193 | 2 | |
| Insertion | Chr1 | 4806996 | 4807004 | 9 | |
| Insertion | Chr10 | 10255192 | 10255202 | 11 | |
| Insertion | Chr10 | 14887839 | 14887839 | 1 | |
| Insertion | Chr12 | 77173 | 77249 | 77 | |
| Insertion | Chr4 | 20973567 | 20973574 | 8 |
No Inversion
No Translocation
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr10 | 5064830 | 5064830 | 0 | LOC_Os10g09390 |
