Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1526-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1526-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 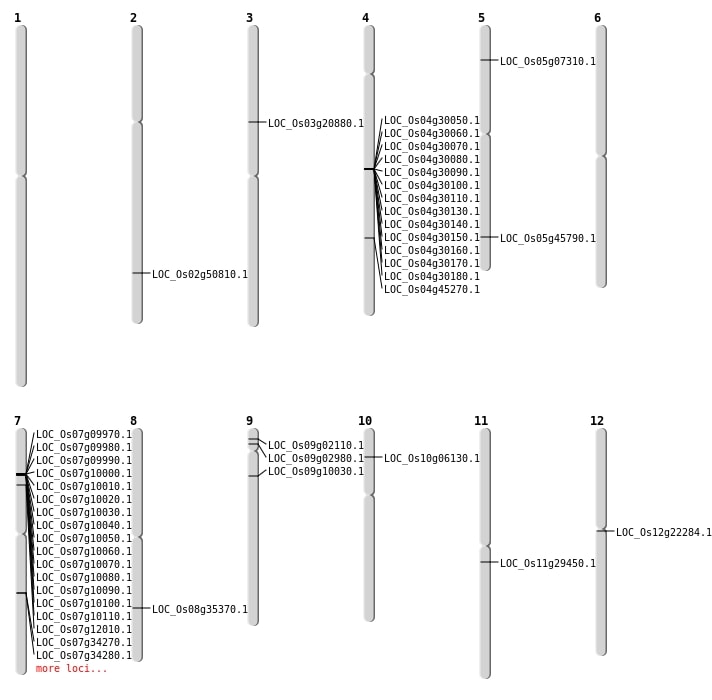
|
| Variant Information |
|---|
Single base substitutions: 23
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21801752 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 2665738 | C-A | HET | INTRON | |
| SBS | Chr1 | 2695879 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 31420043 | A-T | HET | INTRON | |
| SBS | Chr1 | 35008744 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 1281700 | G-T | HET | INTRON | |
| SBS | Chr10 | 1281701 | A-T | HET | INTRON | |
| SBS | Chr10 | 16060305 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 15180057 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 16212293 | A-G | HET | INTRON | |
| SBS | Chr11 | 17428514 | G-A | Homo | INTRON | |
| SBS | Chr12 | 1693482 | C-T | Homo | INTERGENIC | |
| SBS | Chr2 | 13696406 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 34816459 | C-T | Homo | INTERGENIC | |
| SBS | Chr4 | 14879797 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 18158084 | G-A | Homo | INTERGENIC | |
| SBS | Chr5 | 19578950 | A-G | Homo | INTERGENIC | |
| SBS | Chr6 | 2421030 | C-T | Homo | UTR_3_PRIME | |
| SBS | Chr7 | 16194484 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 23153066 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 17432490 | A-T | Homo | INTERGENIC | |
| SBS | Chr9 | 1399392 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g02980 |
| SBS | Chr9 | 5465584 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g10030 |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 791728 | 791738 | 10 | LOC_Os09g02110 |
| Deletion | Chr1 | 2149793 | 2149831 | 38 | |
| Deletion | Chr10 | 3117363 | 3117397 | 34 | LOC_Os10g06130 |
| Deletion | Chr12 | 3666309 | 3666310 | 1 | |
| Deletion | Chr5 | 3889859 | 3889860 | 1 | LOC_Os05g07310 |
| Deletion | Chr11 | 4205693 | 4205702 | 9 | |
| Deletion | Chr7 | 5313001 | 5421000 | 108000 | 15 |
| Deletion | Chr7 | 6671153 | 6671154 | 1 | LOC_Os07g12010 |
| Deletion | Chr9 | 7496982 | 7496987 | 5 | |
| Deletion | Chr10 | 8967538 | 8967541 | 3 | |
| Deletion | Chr3 | 9018043 | 9018044 | 1 | |
| Deletion | Chr1 | 10924623 | 10924629 | 6 | |
| Deletion | Chr5 | 10943421 | 10943424 | 3 | |
| Deletion | Chr3 | 11819256 | 11821526 | 2270 | LOC_Os03g20880 |
| Deletion | Chr12 | 12569136 | 12569137 | 1 | LOC_Os12g22284 |
| Deletion | Chr5 | 12614904 | 12614906 | 2 | |
| Deletion | Chr3 | 12859658 | 12859659 | 1 | |
| Deletion | Chr11 | 17093980 | 17093983 | 3 | LOC_Os11g29450 |
| Deletion | Chr4 | 17918001 | 18014000 | 96000 | 14 |
| Deletion | Chr6 | 19850157 | 19850163 | 6 | |
| Deletion | Chr7 | 20517001 | 20610000 | 93000 | 12 |
| Deletion | Chr7 | 22334539 | 22334540 | 1 | |
| Deletion | Chr6 | 22370521 | 22370605 | 84 | |
| Deletion | Chr4 | 23681154 | 23681155 | 1 | |
| Deletion | Chr7 | 24144354 | 24144355 | 1 | |
| Deletion | Chr8 | 24810669 | 24810673 | 4 | |
| Deletion | Chr4 | 26746742 | 26746749 | 7 | LOC_Os04g45270 |
| Deletion | Chr1 | 27331888 | 27331996 | 108 | |
| Deletion | Chr1 | 29362114 | 29362121 | 7 | |
| Deletion | Chr2 | 31038833 | 31042155 | 3322 | LOC_Os02g50810 |
| Deletion | Chr3 | 36197599 | 36197600 | 1 |
Insertions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 6150338 | 6150338 | 1 | |
| Insertion | Chr2 | 25191437 | 25191502 | 66 | |
| Insertion | Chr3 | 11910334 | 11910335 | 2 | |
| Insertion | Chr4 | 28700438 | 28700438 | 1 | |
| Insertion | Chr4 | 29519598 | 29519598 | 1 | |
| Insertion | Chr4 | 7618518 | 7618521 | 4 | |
| Insertion | Chr5 | 26526871 | 26526945 | 75 | LOC_Os05g45790 |
| Insertion | Chr6 | 9995080 | 9995080 | 1 | |
| Insertion | Chr7 | 7347127 | 7347127 | 1 | |
| Insertion | Chr8 | 22300161 | 22300163 | 3 | LOC_Os08g35370 |
| Insertion | Chr8 | 8652045 | 8652082 | 38 | |
| Insertion | Chr9 | 15067187 | 15067188 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 9208783 | 9334064 | |
| Inversion | Chr8 | 9208804 | 9334070 |
No Translocation
