Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1527-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1527-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 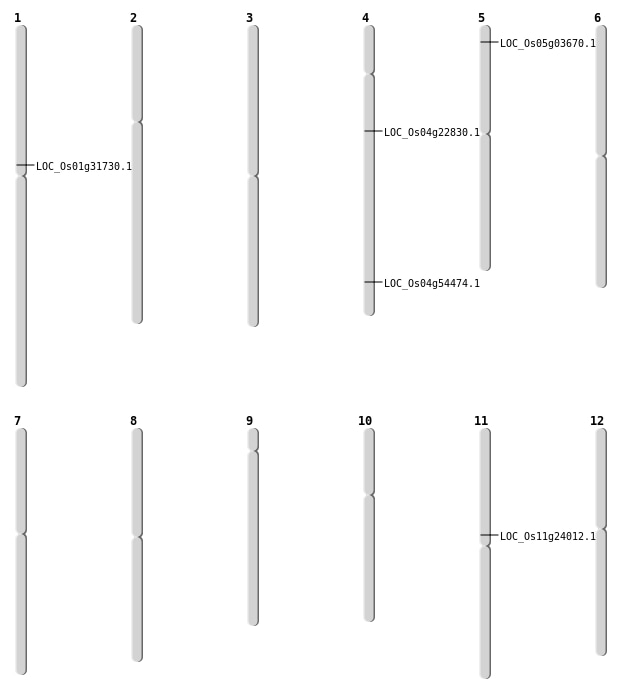
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12243692 | T-A | Homo | INTRON | |
| SBS | Chr1 | 12243709 | T-A | Homo | SPLICE_SITE_REGION | |
| SBS | Chr1 | 16673745 | T-C | Homo | INTERGENIC | |
| SBS | Chr1 | 17364457 | G-A | Homo | SYNONYMOUS_CODING | |
| SBS | Chr1 | 17364458 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g31730 |
| SBS | Chr1 | 28987158 | G-A | Homo | INTERGENIC | |
| SBS | Chr1 | 29752308 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 29752364 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 29975787 | C-A | Homo | INTERGENIC | |
| SBS | Chr1 | 31805080 | C-T | Homo | INTERGENIC | |
| SBS | Chr1 | 5946678 | G-A | Homo | SYNONYMOUS_CODING | |
| SBS | Chr10 | 12824281 | G-A | Homo | INTRON | |
| SBS | Chr10 | 1953673 | C-T | Homo | INTRON | |
| SBS | Chr11 | 13610497 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g24012 |
| SBS | Chr11 | 14494751 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 16559318 | C-A | Homo | INTERGENIC | |
| SBS | Chr12 | 10322774 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 12806022 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 15075708 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 24134808 | T-G | HET | UTR_3_PRIME | |
| SBS | Chr2 | 24145902 | C-T | HET | INTRON | |
| SBS | Chr3 | 19517377 | C-T | Homo | INTRON | |
| SBS | Chr4 | 12929522 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g22830 |
| SBS | Chr4 | 3934835 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 4824733 | G-A | Homo | INTERGENIC | |
| SBS | Chr4 | 5939594 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 1785430 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr5 | 21918842 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 9679524 | A-T | Homo | INTERGENIC | |
| SBS | Chr6 | 9502462 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 13265470 | G-C | HET | INTRON | |
| SBS | Chr7 | 15731244 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 29421832 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 29421833 | A-T | Homo | INTERGENIC | |
| SBS | Chr8 | 11458210 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 1687667 | A-T | HET | INTRON |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 815631 | 815632 | 1 | |
| Deletion | Chr8 | 1006602 | 1006604 | 2 | |
| Deletion | Chr1 | 1304854 | 1304856 | 2 | |
| Deletion | Chr5 | 1307574 | 1307575 | 1 | |
| Deletion | Chr5 | 1590394 | 1590417 | 23 | LOC_Os05g03670 |
| Deletion | Chr1 | 2551382 | 2551390 | 8 | |
| Deletion | Chr8 | 3253114 | 3253116 | 2 | |
| Deletion | Chr8 | 3260332 | 3260333 | 1 | |
| Deletion | Chr1 | 5044718 | 5044719 | 1 | |
| Deletion | Chr10 | 6318377 | 6318397 | 20 | |
| Deletion | Chr10 | 7540885 | 7540897 | 12 | |
| Deletion | Chr2 | 8162433 | 8162444 | 11 | |
| Deletion | Chr11 | 8162436 | 8162438 | 2 | |
| Deletion | Chr1 | 11598262 | 11598263 | 1 | |
| Deletion | Chr12 | 15242197 | 15242259 | 62 | |
| Deletion | Chr7 | 17425121 | 17425122 | 1 | |
| Deletion | Chr3 | 18904340 | 18904342 | 2 | |
| Deletion | Chr6 | 22150106 | 22150107 | 1 | |
| Deletion | Chr7 | 22351212 | 22351216 | 4 | |
| Deletion | Chr4 | 25689574 | 25689601 | 27 | |
| Deletion | Chr8 | 27874539 | 27874540 | 1 | |
| Deletion | Chr6 | 28273145 | 28273146 | 1 | |
| Deletion | Chr11 | 28910326 | 28910337 | 11 | |
| Deletion | Chr4 | 32411436 | 32411441 | 5 | LOC_Os04g54474 |
| Deletion | Chr2 | 34972201 | 34972202 | 1 |
Insertions: 15
No Inversion
No Translocation
