Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1528-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1528-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 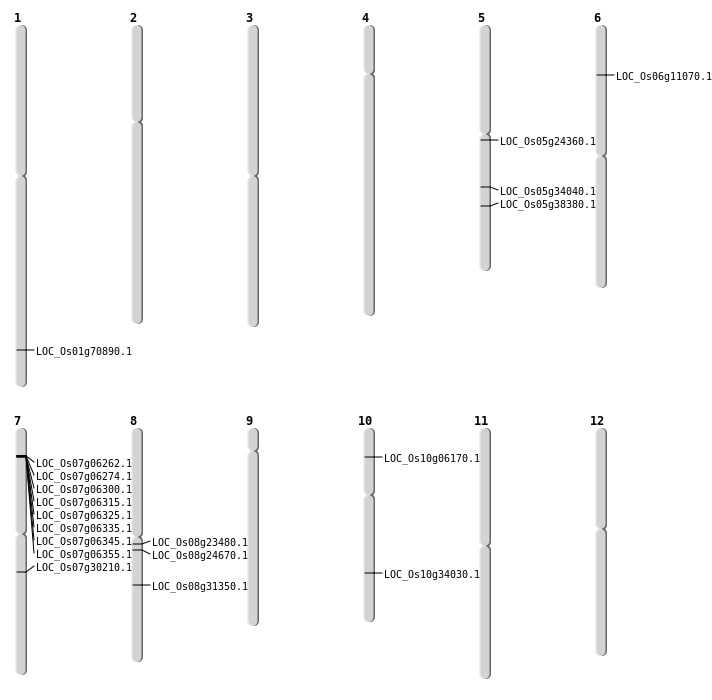
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16969455 | T-G | HET | INTRON | |
| SBS | Chr1 | 23997665 | C-T | Homo | INTERGENIC | |
| SBS | Chr1 | 31931304 | C-T | Homo | INTERGENIC | |
| SBS | Chr1 | 33779982 | G-T | Homo | INTERGENIC | |
| SBS | Chr10 | 10630484 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 18384422 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 19878286 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 3158239 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g06170 |
| SBS | Chr12 | 9781764 | T-A | HET | INTRON | |
| SBS | Chr2 | 29339017 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 896727 | A-T | Homo | INTRON | |
| SBS | Chr3 | 11743012 | C-T | HET | INTRON | |
| SBS | Chr3 | 14364842 | T-C | HET | INTRON | |
| SBS | Chr3 | 30012003 | G-C | HET | INTRON | |
| SBS | Chr3 | 6875624 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 9319195 | A-C | Homo | INTERGENIC | |
| SBS | Chr4 | 17461317 | A-T | Homo | INTRON | |
| SBS | Chr4 | 21825147 | C-T | Homo | INTERGENIC | |
| SBS | Chr4 | 28692908 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 13248569 | C-G | HET | INTRON | |
| SBS | Chr5 | 14087701 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g24360 |
| SBS | Chr5 | 24858333 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 7148610 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 12456513 | C-T | Homo | INTERGENIC | |
| SBS | Chr6 | 12456519 | A-G | Homo | INTERGENIC | |
| SBS | Chr6 | 3671739 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 14217493 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g23480 |
| SBS | Chr8 | 17142987 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 5022225 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 12160655 | C-T | HET | INTERGENIC |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 2216295 | 2216297 | 2 | |
| Deletion | Chr7 | 3042001 | 3098000 | 56000 | 8 |
| Deletion | Chr3 | 3071693 | 3071694 | 1 | |
| Deletion | Chr7 | 3543531 | 3543533 | 2 | |
| Deletion | Chr1 | 3746064 | 3746075 | 11 | |
| Deletion | Chr2 | 4532594 | 4532604 | 10 | |
| Deletion | Chr6 | 5807453 | 5807462 | 9 | LOC_Os06g11070 |
| Deletion | Chr1 | 8275943 | 8275955 | 12 | |
| Deletion | Chr11 | 9375664 | 9375665 | 1 | |
| Deletion | Chr11 | 11896806 | 11896813 | 7 | |
| Deletion | Chr1 | 12555127 | 12555135 | 8 | |
| Deletion | Chr5 | 13285058 | 13285063 | 5 | |
| Deletion | Chr11 | 13914270 | 13914298 | 28 | |
| Deletion | Chr8 | 14909503 | 14909507 | 4 | LOC_Os08g24670 |
| Deletion | Chr1 | 16738962 | 16738973 | 11 | |
| Deletion | Chr5 | 17290662 | 17290663 | 1 | |
| Deletion | Chr3 | 17791463 | 17791466 | 3 | |
| Deletion | Chr10 | 18384427 | 18384428 | 1 | |
| Deletion | Chr9 | 18823867 | 18823873 | 6 | |
| Deletion | Chr4 | 19810475 | 19810485 | 10 | |
| Deletion | Chr2 | 20330631 | 20330638 | 7 | |
| Deletion | Chr8 | 21049773 | 21049777 | 4 | |
| Deletion | Chr9 | 21572614 | 21572615 | 1 | |
| Deletion | Chr4 | 21866646 | 21866652 | 6 | |
| Deletion | Chr1 | 22483885 | 22483910 | 25 | |
| Deletion | Chr5 | 22496701 | 22496706 | 5 | LOC_Os05g38380 |
| Deletion | Chr7 | 23804768 | 23804771 | 3 | |
| Deletion | Chr3 | 23863613 | 23863614 | 1 | |
| Deletion | Chr7 | 25367604 | 25367606 | 2 | |
| Deletion | Chr12 | 27167799 | 27167806 | 7 | |
| Deletion | Chr4 | 30665214 | 30665215 | 1 | |
| Deletion | Chr3 | 30798910 | 30798939 | 29 | |
| Deletion | Chr2 | 31487333 | 31487343 | 10 | |
| Deletion | Chr1 | 41029268 | 41029283 | 15 | LOC_Os01g70890 |
Insertions: 16
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 17848616 | 21727674 | LOC_Os07g30210 |
| Inversion | Chr10 | 17999867 | 18150615 | LOC_Os10g34030 |
| Inversion | Chr10 | 18024714 | 18150620 | LOC_Os10g34030 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 15755884 | Chr5 | 20107683 | LOC_Os05g34040 |
| Translocation | Chr6 | 18092328 | Chr2 | 10168583 | |
| Translocation | Chr6 | 18092332 | Chr2 | 10134136 | |
| Translocation | Chr8 | 19390399 | Chr3 | 19732442 | LOC_Os08g31350 |
