Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1531-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1531-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 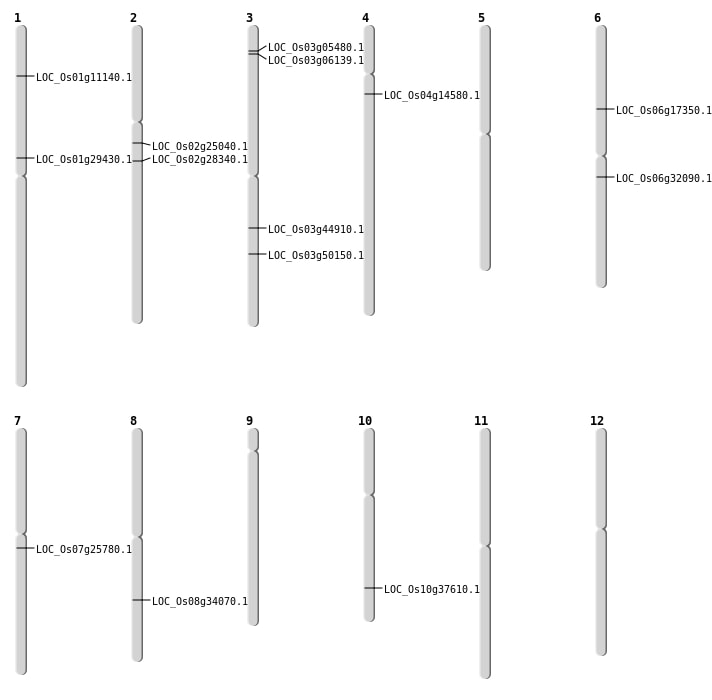
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 18941317 | C-T | Homo | UTR_3_PRIME | |
| SBS | Chr11 | 12185485 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 8676034 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 12561331 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 1898014 | C-T | Homo | INTRON | |
| SBS | Chr12 | 19746825 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 19746826 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 25110381 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 3295040 | T-C | Homo | INTERGENIC | |
| SBS | Chr2 | 31782805 | C-A | HET | INTRON | |
| SBS | Chr2 | 35185984 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 7392008 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 15265725 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 16340082 | T-C | Homo | INTERGENIC | |
| SBS | Chr3 | 20510184 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 21778418 | G-A | HET | INTRON | |
| SBS | Chr3 | 32364484 | C-T | Homo | INTERGENIC | |
| SBS | Chr4 | 27692997 | T-C | Homo | INTRON | |
| SBS | Chr4 | 33801321 | T-C | Homo | INTERGENIC | |
| SBS | Chr4 | 792950 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 19456539 | A-G | HET | INTRON | |
| SBS | Chr6 | 2617627 | C-T | HET | INTRON | |
| SBS | Chr7 | 14428935 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 14810792 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g25780 |
| SBS | Chr7 | 1546949 | T-C | Homo | INTERGENIC | |
| SBS | Chr7 | 15914161 | G-T | Homo | INTRON | |
| SBS | Chr7 | 9728825 | T-C | Homo | INTERGENIC | |
| SBS | Chr8 | 12832522 | C-A | Homo | INTRON | |
| SBS | Chr8 | 248535 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 27300988 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 1072166 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 14478317 | G-A | HET | INTERGENIC |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 5007865 | 5007871 | 6 | |
| Deletion | Chr2 | 5226585 | 5228052 | 1467 | |
| Deletion | Chr1 | 5965564 | 5965565 | 1 | LOC_Os01g11140 |
| Deletion | Chr2 | 6574686 | 6574718 | 32 | |
| Deletion | Chr10 | 7461297 | 7461301 | 4 | |
| Deletion | Chr1 | 7903367 | 7903368 | 1 | |
| Deletion | Chr12 | 9301670 | 9301671 | 1 | |
| Deletion | Chr2 | 14521857 | 14521866 | 9 | LOC_Os02g25040 |
| Deletion | Chr1 | 16495975 | 16495976 | 1 | LOC_Os01g29430 |
| Deletion | Chr5 | 19517060 | 19517076 | 16 | |
| Deletion | Chr7 | 21264426 | 21264464 | 38 | |
| Deletion | Chr4 | 22469679 | 22469680 | 1 | |
| Deletion | Chr11 | 27254299 | 27254300 | 1 | |
| Deletion | Chr3 | 28601199 | 28601201 | 2 | LOC_Os03g50150 |
| Deletion | Chr1 | 29538958 | 29538962 | 4 | |
| Deletion | Chr6 | 29698675 | 29698692 | 17 |
Insertions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 12717709 | 12717709 | 1 | |
| Insertion | Chr11 | 28399052 | 28399053 | 2 | |
| Insertion | Chr12 | 13003863 | 13003863 | 1 | |
| Insertion | Chr2 | 16751019 | 16751021 | 3 | LOC_Os02g28340 |
| Insertion | Chr2 | 25893500 | 25893506 | 7 | |
| Insertion | Chr3 | 25355891 | 25355911 | 21 | LOC_Os03g44910 |
| Insertion | Chr3 | 3839673 | 3839730 | 58 | |
| Insertion | Chr4 | 731102 | 731134 | 33 | |
| Insertion | Chr4 | 8178339 | 8178384 | 46 | LOC_Os04g14580 |
| Insertion | Chr5 | 11573423 | 11573424 | 2 | |
| Insertion | Chr5 | 28560675 | 28560675 | 1 | |
| Insertion | Chr6 | 14142962 | 14142965 | 4 | |
| Insertion | Chr6 | 18674478 | 18674478 | 1 | LOC_Os06g32090 |
| Insertion | Chr6 | 19429525 | 19429584 | 60 | |
| Insertion | Chr7 | 23815210 | 23815211 | 2 | |
| Insertion | Chr7 | 27764597 | 27764676 | 80 | |
| Insertion | Chr8 | 12511620 | 12511622 | 3 | |
| Insertion | Chr8 | 804772 | 804772 | 1 | |
| Insertion | Chr9 | 4900406 | 4900406 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 743177 | 35479073 | |
| Inversion | Chr3 | 2713675 | 3084916 | 2 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 20128805 | Chr3 | 3084990 | 2 |
| Translocation | Chr10 | 20129590 | Chr3 | 2713677 | LOC_Os03g05480 |
| Translocation | Chr10 | 20350302 | Chr6 | 10075406 | |
| Translocation | Chr10 | 20350306 | Chr6 | 10058612 | LOC_Os06g17350 |
| Translocation | Chr8 | 21357466 | Chr6 | 607902 | LOC_Os08g34070 |
