Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1533-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1533-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 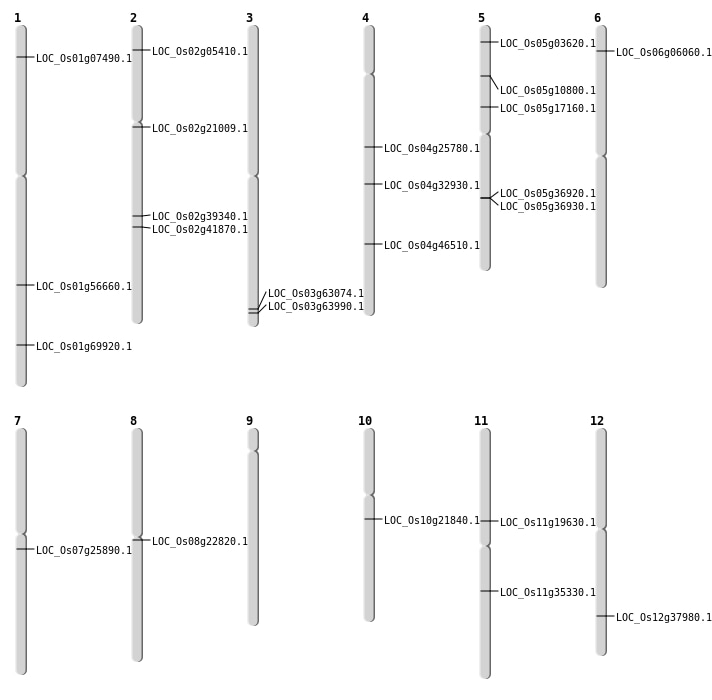
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13393513 | G-A | Homo | INTERGENIC | |
| SBS | Chr1 | 13393515 | G-A | Homo | INTERGENIC | |
| SBS | Chr1 | 22681841 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 22681842 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 26161283 | G-A | Homo | INTERGENIC | |
| SBS | Chr1 | 40292919 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 42067988 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 9838497 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 15587033 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 19533257 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 2041592 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 4562341 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 11313522 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g19630 |
| SBS | Chr11 | 11313524 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g19630 |
| SBS | Chr11 | 25073409 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 11447632 | T-C | Homo | INTERGENIC | |
| SBS | Chr12 | 22490891 | A-G | HET | INTRON | |
| SBS | Chr2 | 17630531 | G-A | HET | INTRON | |
| SBS | Chr2 | 23748269 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g39340 |
| SBS | Chr2 | 35100894 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 27595191 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 18723915 | A-G | Homo | INTERGENIC | |
| SBS | Chr4 | 22975886 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 27586977 | C-A | Homo | STOP_GAINED | LOC_Os04g46510 |
| SBS | Chr4 | 4672318 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 11377491 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 14201319 | C-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr5 | 2356719 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 28070760 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 4201392 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 5154448 | C-A | Homo | INTRON | |
| SBS | Chr5 | 9829249 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g17160 |
| SBS | Chr6 | 20303444 | A-C | Homo | SYNONYMOUS_CODING | |
| SBS | Chr7 | 14402631 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 13723591 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g22820 |
| SBS | Chr9 | 7136797 | T-A | HET | INTRON |
Deletions: 60
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 572358 | 581935 | 9577 | |
| Deletion | Chr4 | 1276016 | 1276026 | 10 | |
| Deletion | Chr9 | 1373073 | 1373086 | 13 | |
| Deletion | Chr5 | 1548432 | 1548664 | 232 | LOC_Os05g03620 |
| Deletion | Chr2 | 2602892 | 2602902 | 10 | LOC_Os02g05410 |
| Deletion | Chr6 | 2789909 | 2789910 | 1 | LOC_Os06g06060 |
| Deletion | Chr4 | 4129140 | 4129141 | 1 | |
| Deletion | Chr5 | 4341307 | 4341308 | 1 | |
| Deletion | Chr11 | 4705153 | 4705155 | 2 | |
| Deletion | Chr10 | 5123859 | 5123860 | 1 | |
| Deletion | Chr6 | 5576576 | 5576611 | 35 | |
| Deletion | Chr5 | 5965664 | 5965871 | 207 | LOC_Os05g10800 |
| Deletion | Chr10 | 6037918 | 6037926 | 8 | |
| Deletion | Chr2 | 6475511 | 6475515 | 4 | |
| Deletion | Chr8 | 7096095 | 7096169 | 74 | |
| Deletion | Chr6 | 8253646 | 8253660 | 14 | |
| Deletion | Chr5 | 9390192 | 9390198 | 6 | |
| Deletion | Chr8 | 9504484 | 9504485 | 1 | |
| Deletion | Chr11 | 10153830 | 10153833 | 3 | |
| Deletion | Chr10 | 11225626 | 11225630 | 4 | LOC_Os10g21840 |
| Deletion | Chr3 | 11386449 | 11386450 | 1 | |
| Deletion | Chr10 | 11591516 | 11591527 | 11 | |
| Deletion | Chr10 | 12529518 | 12529519 | 1 | |
| Deletion | Chr5 | 14383035 | 14383038 | 3 | |
| Deletion | Chr7 | 14867567 | 14867569 | 2 | LOC_Os07g25890 |
| Deletion | Chr4 | 14899188 | 14899189 | 1 | |
| Deletion | Chr4 | 14945840 | 14945847 | 7 | LOC_Os04g25780 |
| Deletion | Chr4 | 15229488 | 15229773 | 285 | |
| Deletion | Chr2 | 15265474 | 15265476 | 2 | |
| Deletion | Chr11 | 15324393 | 15324394 | 1 | |
| Deletion | Chr2 | 15465192 | 15465198 | 6 | |
| Deletion | Chr8 | 15917031 | 15917032 | 1 | |
| Deletion | Chr4 | 15985177 | 15985181 | 4 | |
| Deletion | Chr5 | 16117011 | 16117012 | 1 | |
| Deletion | Chr9 | 16309330 | 16309339 | 9 | |
| Deletion | Chr7 | 16583611 | 16583612 | 1 | |
| Deletion | Chr11 | 16647167 | 16647169 | 2 | |
| Deletion | Chr4 | 17115958 | 17115976 | 18 | |
| Deletion | Chr10 | 17410618 | 17410635 | 17 | |
| Deletion | Chr6 | 17488651 | 17488652 | 1 | |
| Deletion | Chr11 | 17886534 | 17886537 | 3 | |
| Deletion | Chr8 | 18275080 | 18275083 | 3 | |
| Deletion | Chr5 | 18458916 | 18458922 | 6 | |
| Deletion | Chr4 | 19333450 | 19333457 | 7 | |
| Deletion | Chr12 | 20268863 | 20268864 | 1 | |
| Deletion | Chr11 | 20717113 | 20717130 | 17 | LOC_Os11g35330 |
| Deletion | Chr3 | 20731124 | 20731137 | 13 | |
| Deletion | Chr1 | 21110813 | 21110824 | 11 | |
| Deletion | Chr8 | 21549544 | 21549557 | 13 | |
| Deletion | Chr9 | 21843129 | 21843130 | 1 | |
| Deletion | Chr8 | 24653182 | 24653189 | 7 | |
| Deletion | Chr2 | 25167344 | 25167346 | 2 | LOC_Os02g41870 |
| Deletion | Chr8 | 27001411 | 27001416 | 5 | |
| Deletion | Chr3 | 28597939 | 28597940 | 1 | |
| Deletion | Chr4 | 28755108 | 28755109 | 1 | |
| Deletion | Chr1 | 32694761 | 32694762 | 1 | LOC_Os01g56660 |
| Deletion | Chr2 | 33961057 | 33961058 | 1 | |
| Deletion | Chr3 | 35662876 | 35662877 | 1 | LOC_Os03g63074 |
| Deletion | Chr3 | 36167980 | 36167990 | 10 | LOC_Os03g63990 |
| Deletion | Chr1 | 40409455 | 40409458 | 3 | LOC_Os01g69920 |
Insertions: 9
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 21563168 | 21571556 | 2 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 5304674 | Chr10 | 4271318 | |
| Translocation | Chr4 | 19893639 | Chr1 | 3561836 | 2 |
| Translocation | Chr4 | 19893699 | Chr1 | 3559919 | 2 |
| Translocation | Chr12 | 23339555 | Chr2 | 12446081 | 2 |
| Translocation | Chr12 | 23339556 | Chr2 | 12442517 | 2 |
