Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1535-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1535-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 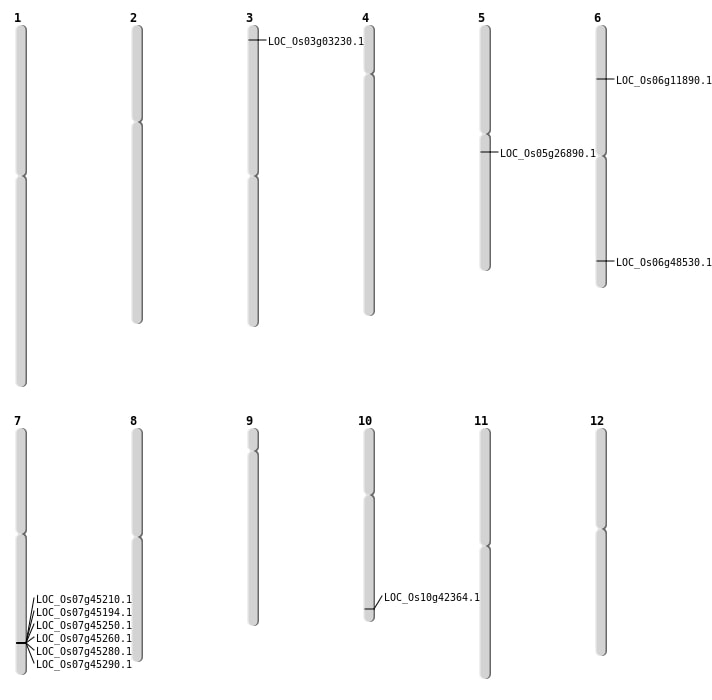
|
| Variant Information |
|---|
Single base substitutions: 26
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15268331 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 5920820 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 13521887 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 21726403 | A-G | Homo | INTRON | |
| SBS | Chr11 | 16463578 | C-T | Homo | INTERGENIC | |
| SBS | Chr12 | 3995526 | A-G | Homo | INTERGENIC | |
| SBS | Chr2 | 17723697 | G-A | HET | INTRON | |
| SBS | Chr2 | 2425258 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 7034771 | G-T | HET | UTR_5_PRIME | |
| SBS | Chr2 | 7034772 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr3 | 28937040 | T-C | HET | INTRON | |
| SBS | Chr3 | 8775183 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 4991656 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 1107683 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 17364138 | G-A | Homo | INTERGENIC | |
| SBS | Chr6 | 29360930 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g48530 |
| SBS | Chr6 | 6083415 | T-C | Homo | SYNONYMOUS_CODING | |
| SBS | Chr6 | 6322916 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g11890 |
| SBS | Chr7 | 14122189 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 23784384 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 23857134 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 27588474 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 6155330 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 18375980 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 18646147 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 6685630 | T-C | HET | INTERGENIC |
Deletions: 38
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1286963 | 1286966 | 3 | |
| Deletion | Chr3 | 1372601 | 1372602 | 1 | LOC_Os03g03230 |
| Deletion | Chr12 | 2349867 | 2349909 | 42 | |
| Deletion | Chr12 | 2624702 | 2624709 | 7 | |
| Deletion | Chr11 | 3963541 | 3963547 | 6 | |
| Deletion | Chr11 | 4173414 | 4173424 | 10 | |
| Deletion | Chr1 | 4442507 | 4442508 | 1 | |
| Deletion | Chr1 | 5184622 | 5184624 | 2 | |
| Deletion | Chr1 | 5780259 | 5780263 | 4 | |
| Deletion | Chr11 | 5785664 | 5785665 | 1 | |
| Deletion | Chr5 | 6539473 | 6539474 | 1 | |
| Deletion | Chr2 | 9019495 | 9019496 | 1 | |
| Deletion | Chr2 | 10414481 | 10414494 | 13 | |
| Deletion | Chr2 | 10700062 | 10700064 | 2 | |
| Deletion | Chr7 | 11644406 | 11644409 | 3 | |
| Deletion | Chr5 | 12129406 | 12129430 | 24 | |
| Deletion | Chr8 | 13136268 | 13136270 | 2 | |
| Deletion | Chr1 | 14824729 | 14824730 | 1 | |
| Deletion | Chr7 | 15319924 | 15319925 | 1 | |
| Deletion | Chr9 | 15957609 | 15957618 | 9 | |
| Deletion | Chr12 | 17707867 | 17707868 | 1 | |
| Deletion | Chr8 | 17783074 | 17783084 | 10 | |
| Deletion | Chr7 | 17801947 | 17801948 | 1 | |
| Deletion | Chr12 | 17838983 | 17838993 | 10 | |
| Deletion | Chr7 | 19130278 | 19130298 | 20 | |
| Deletion | Chr1 | 22097925 | 22097927 | 2 | |
| Deletion | Chr9 | 22395283 | 22395284 | 1 | |
| Deletion | Chr10 | 22809696 | 22809697 | 1 | LOC_Os10g42364 |
| Deletion | Chr10 | 22965992 | 22965997 | 5 | |
| Deletion | Chr8 | 23758830 | 23758832 | 2 | |
| Deletion | Chr5 | 25415392 | 25415411 | 19 | |
| Deletion | Chr5 | 25614709 | 25614710 | 1 | |
| Deletion | Chr5 | 26575111 | 26575121 | 10 | |
| Deletion | Chr7 | 26985001 | 27022000 | 37000 | 6 |
| Deletion | Chr11 | 28439796 | 28439956 | 160 | |
| Deletion | Chr6 | 28639789 | 28639790 | 1 | |
| Deletion | Chr1 | 30141712 | 30141716 | 4 | |
| Deletion | Chr4 | 31521778 | 31521779 | 1 |
Insertions: 10
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 12688119 | 15611997 | LOC_Os05g26890 |
| Inversion | Chr5 | 15612000 | 15858304 | LOC_Os05g26890 |
No Translocation
