Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1537-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1537-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 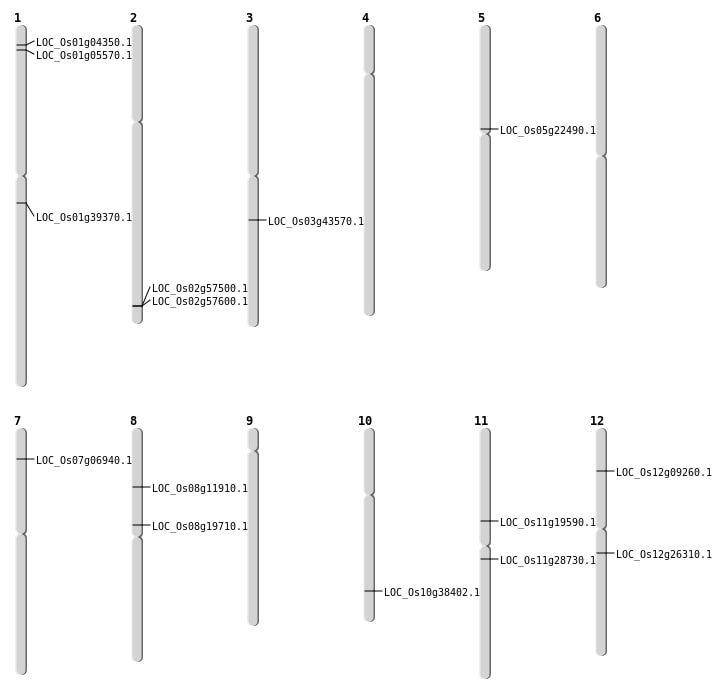
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1926552 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 22158485 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g39370 |
| SBS | Chr1 | 2661195 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g05570 |
| SBS | Chr1 | 38649481 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 38657749 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 4985646 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 9534481 | C-T | HET | INTRON | |
| SBS | Chr10 | 20519626 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g38402 |
| SBS | Chr10 | 797022 | G-A | HET | INTRON | |
| SBS | Chr11 | 11292595 | C-T | Homo | STOP_GAINED | LOC_Os11g19590 |
| SBS | Chr11 | 11933705 | C-A | Homo | INTERGENIC | |
| SBS | Chr11 | 16620160 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g28730 |
| SBS | Chr11 | 16620161 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g28730 |
| SBS | Chr11 | 18589469 | C-A | Homo | INTERGENIC | |
| SBS | Chr11 | 26298561 | G-A | Homo | INTERGENIC | |
| SBS | Chr11 | 341418 | T-A | HET | UTR_5_PRIME | |
| SBS | Chr12 | 11599392 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 15353133 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g26310 |
| SBS | Chr12 | 22227870 | G-A | Homo | INTERGENIC | |
| SBS | Chr12 | 2832304 | A-T | Homo | INTERGENIC | |
| SBS | Chr12 | 4853612 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g09260 |
| SBS | Chr2 | 1192660 | G-C | Homo | INTERGENIC | |
| SBS | Chr2 | 13502023 | T-C | HET | INTRON | |
| SBS | Chr2 | 22671834 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 28902792 | G-A | Homo | INTERGENIC | |
| SBS | Chr3 | 119979 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 15267826 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 23616277 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 29567279 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 35253185 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 9569509 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 9813894 | C-T | HET | INTRON | |
| SBS | Chr4 | 20076997 | G-A | Homo | INTERGENIC | |
| SBS | Chr5 | 6201314 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 6966098 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 20546556 | G-A | Homo | INTERGENIC | |
| SBS | Chr6 | 25994070 | A-G | HET | INTRON | |
| SBS | Chr6 | 27933880 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 21889425 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 3396541 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g06940 |
| SBS | Chr7 | 5259964 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 17949713 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 6981724 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g11910 |
| SBS | Chr9 | 3128637 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 7950153 | T-C | Homo | INTERGENIC |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1941918 | 1941920 | 2 | LOC_Os01g04350 |
| Deletion | Chr12 | 3649302 | 3649303 | 1 | |
| Deletion | Chr3 | 6167704 | 6167711 | 7 | |
| Deletion | Chr8 | 6565272 | 6565289 | 17 | |
| Deletion | Chr10 | 8328910 | 8328911 | 1 | |
| Deletion | Chr6 | 10247645 | 10247646 | 1 | |
| Deletion | Chr1 | 10730362 | 10730364 | 2 | |
| Deletion | Chr1 | 11141217 | 11141218 | 1 | |
| Deletion | Chr8 | 11803673 | 11803677 | 4 | LOC_Os08g19710 |
| Deletion | Chr5 | 12773852 | 12773863 | 11 | LOC_Os05g22490 |
| Deletion | Chr10 | 12996711 | 12996713 | 2 | |
| Deletion | Chr5 | 13221905 | 13221908 | 3 | |
| Deletion | Chr7 | 14362925 | 14362926 | 1 | |
| Deletion | Chr2 | 16463751 | 16463803 | 52 | |
| Deletion | Chr1 | 17772494 | 17772505 | 11 | |
| Deletion | Chr9 | 18140443 | 18140444 | 1 | |
| Deletion | Chr6 | 21161361 | 21161362 | 1 | |
| Deletion | Chr3 | 24341702 | 24341714 | 12 | LOC_Os03g43570 |
| Deletion | Chr3 | 24603818 | 24603819 | 1 | |
| Deletion | Chr2 | 25714198 | 25714199 | 1 | |
| Deletion | Chr8 | 26847964 | 26847965 | 1 | |
| Deletion | Chr2 | 27312590 | 27312600 | 10 | |
| Deletion | Chr12 | 27374058 | 27374063 | 5 | |
| Deletion | Chr5 | 28513700 | 28513702 | 2 | |
| Deletion | Chr4 | 28781127 | 28781132 | 5 | |
| Deletion | Chr4 | 29402058 | 29402059 | 1 | |
| Deletion | Chr3 | 34211157 | 34211158 | 1 | |
| Deletion | Chr2 | 35233904 | 35233908 | 4 | LOC_Os02g57500 |
| Deletion | Chr2 | 35277013 | 35277020 | 7 | LOC_Os02g57600 |
| Deletion | Chr1 | 38289371 | 38289372 | 1 | |
| Deletion | Chr1 | 38900216 | 38900234 | 18 | |
| Deletion | Chr1 | 42517568 | 42517569 | 1 |
Insertions: 13
No Inversion
No Translocation
