Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1541-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1541-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 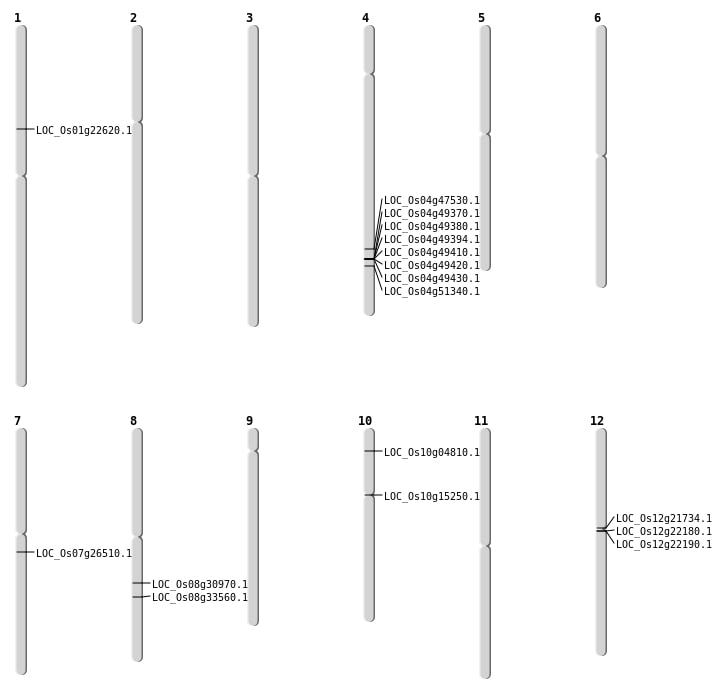
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22511530 | G-A | Homo | INTERGENIC | |
| SBS | Chr1 | 23827064 | C-A | Homo | INTRON | |
| SBS | Chr10 | 1842477 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 8033174 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g15250 |
| SBS | Chr10 | 885719 | C-T | HET | INTRON | |
| SBS | Chr11 | 16467384 | G-A | Homo | INTERGENIC | |
| SBS | Chr11 | 24910820 | T-A | Homo | INTERGENIC | |
| SBS | Chr11 | 25595954 | T-C | Homo | INTERGENIC | |
| SBS | Chr12 | 21916084 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 9896578 | T-A | HET | INTRON | |
| SBS | Chr3 | 16142843 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 24160224 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 26407093 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 36350638 | A-T | HET | INTRON | |
| SBS | Chr3 | 7641587 | C-T | Homo | INTRON | |
| SBS | Chr4 | 23956039 | A-T | HET | INTRON | |
| SBS | Chr4 | 26386450 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 25080659 | G-T | Homo | INTERGENIC | |
| SBS | Chr7 | 25545636 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 13839330 | G-A | Homo | INTERGENIC | |
| SBS | Chr8 | 27075645 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 27943503 | A-T | Homo | INTRON | |
| SBS | Chr8 | 503670 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 6377471 | G-A | Homo | INTERGENIC | |
| SBS | Chr8 | 7076823 | G-A | Homo | INTERGENIC | |
| SBS | Chr9 | 2189299 | G-A | Homo | INTERGENIC | |
| SBS | Chr9 | 3376232 | C-A | Homo | INTERGENIC |
Deletions: 50
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1312208 | 1312219 | 11 | |
| Deletion | Chr4 | 1882356 | 1882358 | 2 | |
| Deletion | Chr10 | 2319660 | 2330605 | 10945 | LOC_Os10g04810 |
| Deletion | Chr10 | 3425642 | 3425661 | 19 | |
| Deletion | Chr6 | 3673815 | 3673817 | 2 | |
| Deletion | Chr11 | 3881664 | 3881666 | 2 | |
| Deletion | Chr11 | 3881665 | 3881667 | 2 | |
| Deletion | Chr10 | 4104700 | 4104705 | 5 | |
| Deletion | Chr6 | 6081072 | 6081082 | 10 | |
| Deletion | Chr9 | 6207424 | 6207425 | 1 | |
| Deletion | Chr2 | 6530910 | 6530911 | 1 | |
| Deletion | Chr12 | 6627473 | 6627564 | 91 | |
| Deletion | Chr11 | 6915421 | 6915422 | 1 | |
| Deletion | Chr10 | 7505244 | 7505245 | 1 | |
| Deletion | Chr2 | 8973662 | 8973663 | 1 | |
| Deletion | Chr12 | 10293452 | 10293453 | 1 | |
| Deletion | Chr12 | 10612875 | 10612884 | 9 | |
| Deletion | Chr11 | 10844967 | 10844968 | 1 | |
| Deletion | Chr1 | 11268215 | 11268219 | 4 | |
| Deletion | Chr5 | 11347624 | 11347626 | 2 | |
| Deletion | Chr12 | 11856301 | 11856302 | 1 | |
| Deletion | Chr7 | 12177312 | 12177313 | 1 | |
| Deletion | Chr12 | 12221941 | 12221947 | 6 | LOC_Os12g21734 |
| Deletion | Chr7 | 12227862 | 12256223 | 28361 | |
| Deletion | Chr12 | 12487882 | 12487889 | 7 | |
| Deletion | Chr12 | 12494142 | 12509483 | 15341 | 2 |
| Deletion | Chr10 | 12752663 | 12752665 | 2 | |
| Deletion | Chr8 | 13355259 | 13355263 | 4 | |
| Deletion | Chr11 | 14213465 | 14213466 | 1 | |
| Deletion | Chr7 | 14905403 | 14905413 | 10 | |
| Deletion | Chr7 | 18305838 | 18305839 | 1 | |
| Deletion | Chr2 | 19065100 | 19065122 | 22 | |
| Deletion | Chr8 | 19119670 | 19119676 | 6 | LOC_Os08g30970 |
| Deletion | Chr12 | 19598429 | 19598430 | 1 | |
| Deletion | Chr2 | 20778670 | 20778687 | 17 | |
| Deletion | Chr8 | 20953281 | 20953282 | 1 | LOC_Os08g33560 |
| Deletion | Chr4 | 22505150 | 22505196 | 46 | |
| Deletion | Chr2 | 22891487 | 22891516 | 29 | |
| Deletion | Chr6 | 23944963 | 23945000 | 37 | |
| Deletion | Chr12 | 25964225 | 25964226 | 1 | |
| Deletion | Chr12 | 26541131 | 26541132 | 1 | |
| Deletion | Chr4 | 27274503 | 27274505 | 2 | |
| Deletion | Chr4 | 27746370 | 27746371 | 1 | |
| Deletion | Chr4 | 28192117 | 28192125 | 8 | LOC_Os04g47530 |
| Deletion | Chr4 | 29360375 | 29360400 | 25 | |
| Deletion | Chr4 | 29465623 | 29494558 | 28935 | 6 |
| Deletion | Chr4 | 30404776 | 30405960 | 1184 | LOC_Os04g51340 |
| Deletion | Chr1 | 31040553 | 31040584 | 31 | |
| Deletion | Chr4 | 31426665 | 31426667 | 2 | |
| Deletion | Chr3 | 34795586 | 34795598 | 12 |
Insertions: 14
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 8091563 | 8501183 | |
| Inversion | Chr7 | 15278713 | 17812182 | LOC_Os07g26510 |
| Inversion | Chr7 | 15278790 | 17812187 | LOC_Os07g26510 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 12494150 | Chr1 | 12724105 | LOC_Os01g22620 |
| Translocation | Chr12 | 12506911 | Chr1 | 12724091 | LOC_Os01g22620 |
| Translocation | Chr12 | 12506913 | Chr3 | 34705298 | LOC_Os03g61080 |
| Translocation | Chr12 | 12509481 | Chr3 | 34705249 | LOC_Os03g61080 |
| Translocation | Chr6 | 14068405 | Chr4 | 23938460 | |
| Translocation | Chr9 | 22372279 | Chr3 | 34870347 | LOC_Os09g38960 |
| Translocation | Chr4 | 30404805 | Chr1 | 28933706 | |
| Translocation | Chr4 | 30405949 | Chr1 | 28933668 | LOC_Os04g51340 |
