Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1548-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1548-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 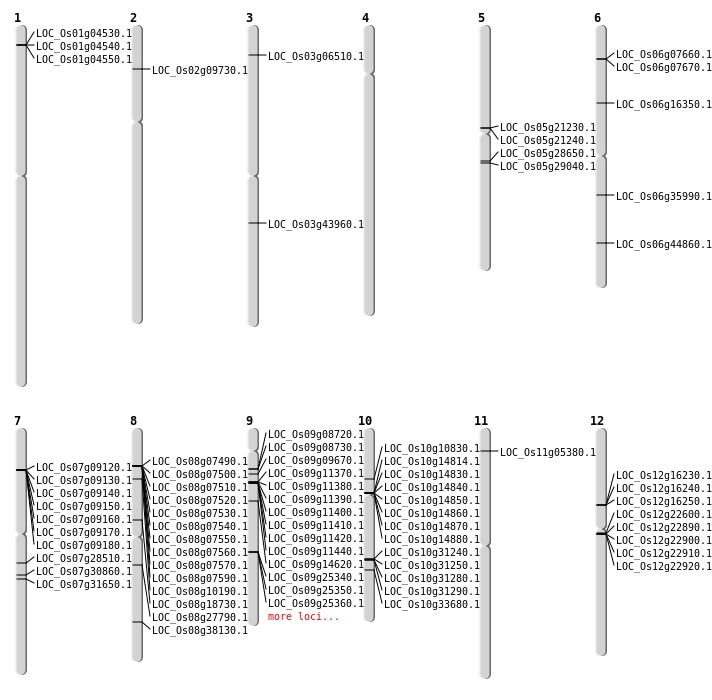
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 36864959 | C-A | HET | ||
| SBS | Chr10 | 11112229 | G-T | Homo | ||
| SBS | Chr10 | 11112230 | A-T | Homo | ||
| SBS | Chr10 | 17762673 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g33680 |
| SBS | Chr10 | 5994912 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g10830 |
| SBS | Chr2 | 16695843 | A-G | HET | ||
| SBS | Chr2 | 28633238 | A-G | Homo | ||
| SBS | Chr2 | 533743 | C-T | Homo | ||
| SBS | Chr2 | 5621033 | G-A | Homo | ||
| SBS | Chr2 | 9017589 | A-C | HET | ||
| SBS | Chr3 | 12716948 | C-A | HET | ||
| SBS | Chr3 | 30842275 | G-A | HET | ||
| SBS | Chr3 | 3268638 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g06510 |
| SBS | Chr4 | 24944748 | C-G | HET | ||
| SBS | Chr4 | 4591859 | G-A | Homo | ||
| SBS | Chr5 | 8799907 | A-G | Homo | ||
| SBS | Chr6 | 11084304 | G-A | HET | ||
| SBS | Chr6 | 15181722 | A-G | HET | ||
| SBS | Chr6 | 25053167 | T-A | HET | ||
| SBS | Chr6 | 25053168 | T-A | HET | ||
| SBS | Chr6 | 28059040 | G-A | HET | ||
| SBS | Chr6 | 5789572 | T-C | HET | ||
| SBS | Chr7 | 16686784 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g28510 |
| SBS | Chr8 | 16927514 | T-C | Homo | START_LOST | LOC_Os08g27790 |
| SBS | Chr8 | 20469216 | A-G | Homo | ||
| SBS | Chr8 | 27063218 | G-A | HET | ||
| SBS | Chr9 | 15830749 | G-A | HET | ||
| SBS | Chr9 | 5215558 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g09670 |
| SBS | Chr9 | 5997905 | G-A | Homo | ||
| SBS | Chr9 | 5997906 | C-T | Homo | ||
| SBS | Chr9 | 8672209 | C-T | Homo | ||
| SBS | Chr9 | 8672210 | C-T | Homo | STOP_GAINED | LOC_Os09g14620 |
Deletions: 41
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 480762 | 480763 | 1 | |
| Deletion | Chr5 | 1564874 | 1564884 | 10 | |
| Deletion | Chr1 | 2019001 | 2037000 | 17999 | 3 |
| Deletion | Chr11 | 2390001 | 2412000 | 21999 | LOC_Os11g05380 |
| Deletion | Chr6 | 3706001 | 3721000 | 14999 | 2 |
| Deletion | Chr6 | 4060614 | 4060621 | 7 | |
| Deletion | Chr8 | 4205001 | 4266000 | 60999 | 10 |
| Deletion | Chr9 | 4587001 | 4603000 | 15999 | 2 |
| Deletion | Chr7 | 4765001 | 4810000 | 44999 | 7 |
| Deletion | Chr2 | 5005001 | 5018000 | 12999 | LOC_Os02g09730 |
| Deletion | Chr8 | 5910928 | 5910938 | 10 | LOC_Os08g10190 |
| Deletion | Chr9 | 5932770 | 5932779 | 9 | |
| Deletion | Chr9 | 6321001 | 6369000 | 47999 | 7 |
| Deletion | Chr2 | 6333331 | 6333332 | 1 | |
| Deletion | Chr10 | 7116559 | 7116562 | 3 | |
| Deletion | Chr10 | 7766001 | 7785000 | 18999 | 7 |
| Deletion | Chr12 | 9278001 | 9294000 | 15999 | 4 |
| Deletion | Chr1 | 9382197 | 9382198 | 1 | |
| Deletion | Chr10 | 9792773 | 9792776 | 3 | |
| Deletion | Chr11 | 10862814 | 10862818 | 4 | |
| Deletion | Chr8 | 11178088 | 11179083 | 995 | LOC_Os08g18730 |
| Deletion | Chr5 | 12550001 | 12577000 | 26999 | 2 |
| Deletion | Chr12 | 12750001 | 12770000 | 19999 | LOC_Os12g22600 |
| Deletion | Chr12 | 12920001 | 12952000 | 31999 | 4 |
| Deletion | Chr9 | 15170001 | 15202000 | 31999 | 3 |
| Deletion | Chr2 | 15442153 | 15442154 | 1 | |
| Deletion | Chr10 | 16383001 | 16416000 | 32999 | 4 |
| Deletion | Chr12 | 16667842 | 16667845 | 3 | |
| Deletion | Chr5 | 16791001 | 16813000 | 21999 | LOC_Os05g28650 |
| Deletion | Chr5 | 17023001 | 17047000 | 23999 | LOC_Os05g29040 |
| Deletion | Chr9 | 17710001 | 17729000 | 18999 | |
| Deletion | Chr2 | 20406461 | 20406484 | 23 | |
| Deletion | Chr6 | 21019001 | 21033000 | 13999 | LOC_Os06g35990 |
| Deletion | Chr9 | 21971001 | 21989000 | 17999 | 2 |
| Deletion | Chr12 | 22829315 | 22829316 | 1 | |
| Deletion | Chr8 | 24158590 | 24158592 | 2 | LOC_Os08g38130 |
| Deletion | Chr3 | 24667001 | 24693000 | 25999 | LOC_Os03g43960 |
| Deletion | Chr12 | 24882844 | 24882849 | 5 | |
| Deletion | Chr6 | 27099001 | 27115000 | 15999 | LOC_Os06g44860 |
| Deletion | Chr2 | 32029286 | 32029293 | 7 | |
| Deletion | Chr1 | 35830526 | 35830528 | 2 |
Insertions: 5
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 1885772 | 1943612 | |
| Inversion | Chr1 | 1885803 | 1943635 | |
| Inversion | Chr6 | 9327150 | 9327436 | LOC_Os06g16350 |
| Inversion | Chr7 | 18266889 | 18805750 | 2 |
| Inversion | Chr7 | 18266892 | 18805751 | 2 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 13891313 | Chr3 | 23252557 | |
| Translocation | Chr6 | 22337528 | Chr3 | 30491781 | |
| Translocation | Chr12 | 25908088 | Chr8 | 23687029 |
