Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1549-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1549-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 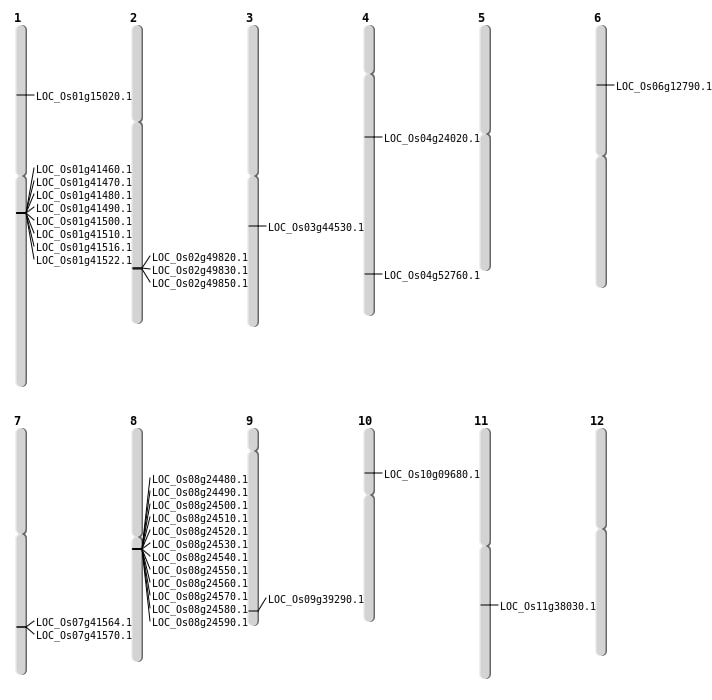
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 29191046 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 35223220 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 43196110 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 8426231 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g15020 |
| SBS | Chr10 | 4927335 | G-C | Homo | INTERGENIC | |
| SBS | Chr11 | 18084954 | G-T | Homo | INTERGENIC | |
| SBS | Chr11 | 25589777 | A-T | Homo | INTERGENIC | |
| SBS | Chr12 | 25393253 | C-T | Homo | INTRON | |
| SBS | Chr12 | 26396582 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 9904304 | A-T | Homo | INTERGENIC | |
| SBS | Chr12 | 9904305 | G-T | Homo | INTERGENIC | |
| SBS | Chr2 | 30468127 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g49850 |
| SBS | Chr2 | 35222143 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr2 | 35533675 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 24714852 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 10439718 | T-A | HET | INTRON | |
| SBS | Chr5 | 19237453 | A-T | HET | INTRON | |
| SBS | Chr5 | 21972921 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 6450756 | C-T | Homo | INTRON | |
| SBS | Chr6 | 1171966 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 2216350 | G-A | Homo | INTERGENIC | |
| SBS | Chr6 | 27599958 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 1835275 | G-C | HET | INTRON | |
| SBS | Chr7 | 28122215 | C-A | HET | INTRON | |
| SBS | Chr7 | 28122220 | C-A | HET | INTRON | |
| SBS | Chr8 | 12696041 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 5620683 | A-C | HET | INTERGENIC |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 327238 | 327239 | 1 | |
| Deletion | Chr12 | 702000 | 702001 | 1 | |
| Deletion | Chr3 | 736012 | 736013 | 1 | |
| Deletion | Chr5 | 907265 | 907266 | 1 | |
| Deletion | Chr2 | 2061683 | 2061695 | 12 | |
| Deletion | Chr4 | 2251327 | 2251332 | 5 | |
| Deletion | Chr2 | 3536991 | 3537001 | 10 | |
| Deletion | Chr4 | 4962714 | 4962727 | 13 | |
| Deletion | Chr10 | 5234255 | 5234260 | 5 | LOC_Os10g09680 |
| Deletion | Chr6 | 6030924 | 6030925 | 1 | |
| Deletion | Chr1 | 6442784 | 6442785 | 1 | |
| Deletion | Chr11 | 6460383 | 6460384 | 1 | |
| Deletion | Chr8 | 8140747 | 8140755 | 8 | |
| Deletion | Chr11 | 9100392 | 9100397 | 5 | |
| Deletion | Chr11 | 12639267 | 12639268 | 1 | |
| Deletion | Chr4 | 13726107 | 13726108 | 1 | LOC_Os04g24020 |
| Deletion | Chr8 | 14789001 | 14875000 | 86000 | 12 |
| Deletion | Chr4 | 16453045 | 16453047 | 2 | |
| Deletion | Chr5 | 21280811 | 21280812 | 1 | |
| Deletion | Chr11 | 22562387 | 22562389 | 2 | LOC_Os11g38030 |
| Deletion | Chr2 | 22728873 | 22728878 | 5 | |
| Deletion | Chr1 | 23467001 | 23509000 | 42000 | 8 |
| Deletion | Chr11 | 24838616 | 24838623 | 7 | |
| Deletion | Chr7 | 24915260 | 24915267 | 7 | 2 |
| Deletion | Chr3 | 25063165 | 25063172 | 7 | LOC_Os03g44530 |
| Deletion | Chr4 | 25910345 | 25910346 | 1 | |
| Deletion | Chr4 | 27823441 | 27823443 | 2 | |
| Deletion | Chr1 | 30089588 | 30089589 | 1 | |
| Deletion | Chr2 | 30448361 | 30448381 | 20 | 2 |
| Deletion | Chr4 | 31413139 | 31413143 | 4 | LOC_Os04g52760 |
| Deletion | Chr3 | 31582364 | 31582366 | 2 | |
| Deletion | Chr4 | 32111842 | 32111843 | 1 | |
| Deletion | Chr1 | 37704272 | 37704278 | 6 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 30476935 | 30476935 | 1 | |
| Insertion | Chr12 | 10662679 | 10662680 | 2 | |
| Insertion | Chr12 | 22116757 | 22116766 | 10 | |
| Insertion | Chr3 | 36410239 | 36410240 | 2 | |
| Insertion | Chr4 | 3191644 | 3191645 | 2 | |
| Insertion | Chr4 | 9449367 | 9449367 | 1 | |
| Insertion | Chr6 | 6992556 | 6992556 | 1 | LOC_Os06g12790 |
| Insertion | Chr9 | 22585868 | 22585868 | 1 | LOC_Os09g39290 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 3713975 | 12461578 |
No Translocation
