Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1553-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1553-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 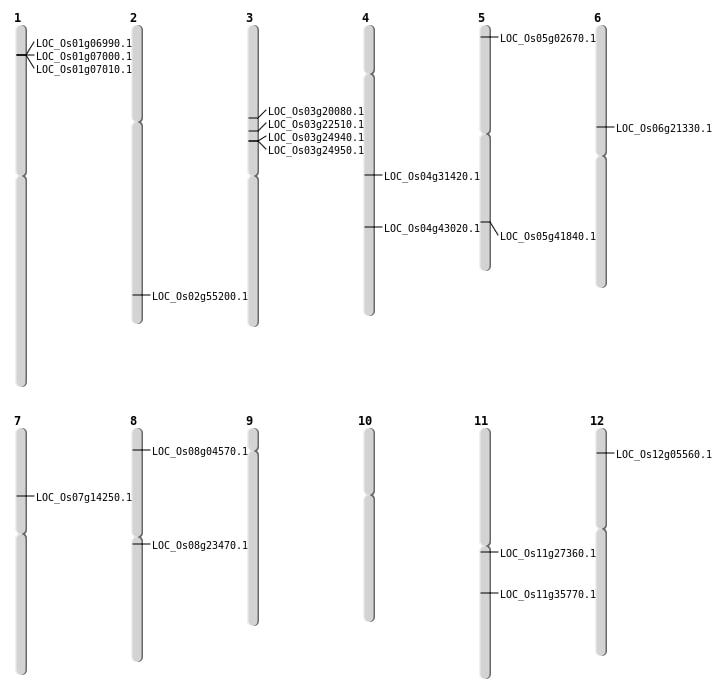
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10512649 | C-T | HET | INTRON | |
| SBS | Chr1 | 5448932 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 13235750 | A-G | HET | INTRON | |
| SBS | Chr11 | 20999563 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g35770 |
| SBS | Chr12 | 24263640 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 2551406 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g05560 |
| SBS | Chr2 | 25064353 | G-T | Homo | INTERGENIC | |
| SBS | Chr2 | 25064354 | G-T | Homo | INTERGENIC | |
| SBS | Chr2 | 29342737 | A-C | HET | INTRON | |
| SBS | Chr2 | 30651489 | A-T | Homo | INTRON | |
| SBS | Chr3 | 10183167 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 10276577 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 11122753 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 33127137 | C-A | Homo | INTERGENIC | |
| SBS | Chr4 | 1605980 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 19618875 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 22328260 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 26663599 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 26666872 | G-T | HET | INTRON | |
| SBS | Chr4 | 26677187 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 29050388 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 940477 | C-A | HET | START_GAINED | LOC_Os05g02670 |
| SBS | Chr6 | 12318935 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g21330 |
| SBS | Chr6 | 27601389 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr7 | 18984209 | A-T | Homo | INTERGENIC | |
| SBS | Chr7 | 9487130 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 18541250 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 2280958 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g04570 |
| SBS | Chr9 | 17474300 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 21155288 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 5123448 | G-A | HET | INTERGENIC |
Deletions: 39
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1413308 | 1413310 | 2 | |
| Deletion | Chr1 | 1824244 | 1824251 | 7 | |
| Deletion | Chr1 | 2659930 | 2659931 | 1 | |
| Deletion | Chr1 | 3297909 | 3303582 | 5673 | 3 |
| Deletion | Chr5 | 3976896 | 3977024 | 128 | |
| Deletion | Chr9 | 5181027 | 5181036 | 9 | |
| Deletion | Chr11 | 7044129 | 7044130 | 1 | |
| Deletion | Chr7 | 7253064 | 7253065 | 1 | |
| Deletion | Chr7 | 8120438 | 8120493 | 55 | LOC_Os07g14250 |
| Deletion | Chr3 | 9793778 | 9793795 | 17 | |
| Deletion | Chr3 | 11314341 | 11314349 | 8 | LOC_Os03g20080 |
| Deletion | Chr3 | 11487831 | 11487874 | 43 | |
| Deletion | Chr1 | 11800678 | 11800683 | 5 | |
| Deletion | Chr7 | 12167574 | 12167575 | 1 | |
| Deletion | Chr8 | 12219726 | 12219727 | 1 | |
| Deletion | Chr3 | 13415126 | 13415127 | 1 | |
| Deletion | Chr3 | 14212001 | 14247000 | 35000 | 2 |
| Deletion | Chr8 | 14213281 | 14213284 | 3 | LOC_Os08g23470 |
| Deletion | Chr1 | 15014398 | 15014400 | 2 | |
| Deletion | Chr11 | 15741679 | 15741681 | 2 | LOC_Os11g27360 |
| Deletion | Chr12 | 17381962 | 17381964 | 2 | |
| Deletion | Chr1 | 18437591 | 18437592 | 1 | |
| Deletion | Chr4 | 18795081 | 18795084 | 3 | LOC_Os04g31420 |
| Deletion | Chr4 | 19617817 | 19617830 | 13 | |
| Deletion | Chr1 | 20675783 | 20675784 | 1 | |
| Deletion | Chr2 | 20822105 | 20822124 | 19 | |
| Deletion | Chr7 | 22427443 | 22427450 | 7 | |
| Deletion | Chr3 | 23099209 | 23099213 | 4 | |
| Deletion | Chr11 | 23560430 | 23560434 | 4 | |
| Deletion | Chr5 | 23619505 | 23619524 | 19 | |
| Deletion | Chr8 | 23806979 | 23806981 | 2 | |
| Deletion | Chr5 | 24504731 | 24504740 | 9 | LOC_Os05g41840 |
| Deletion | Chr4 | 25456844 | 25456846 | 2 | LOC_Os04g43020 |
| Deletion | Chr2 | 25610893 | 25610894 | 1 | |
| Deletion | Chr4 | 25691245 | 25691246 | 1 | |
| Deletion | Chr8 | 25786856 | 25786860 | 4 | |
| Deletion | Chr5 | 28083260 | 28083261 | 1 | |
| Deletion | Chr2 | 34540458 | 34540466 | 8 | |
| Deletion | Chr1 | 37349333 | 37349334 | 1 |
Insertions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 18727838 | 18727838 | 1 | |
| Insertion | Chr1 | 23885835 | 23885835 | 1 | |
| Insertion | Chr1 | 26513763 | 26513764 | 2 | |
| Insertion | Chr11 | 2043359 | 2043364 | 6 | |
| Insertion | Chr2 | 19232817 | 19232818 | 2 | |
| Insertion | Chr2 | 33811683 | 33811751 | 69 | LOC_Os02g55200 |
| Insertion | Chr3 | 12930149 | 12930150 | 2 | LOC_Os03g22510 |
| Insertion | Chr3 | 30330711 | 30330711 | 1 | |
| Insertion | Chr6 | 10435328 | 10435359 | 32 | |
| Insertion | Chr6 | 1338435 | 1338462 | 28 | |
| Insertion | Chr7 | 18835045 | 18835045 | 1 | |
| Insertion | Chr7 | 23041080 | 23041080 | 1 | |
| Insertion | Chr9 | 10580630 | 10580631 | 2 | |
| Insertion | Chr9 | 8986295 | 8986297 | 3 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 5196036 | 5680508 |
No Translocation
