Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1566-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1566-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 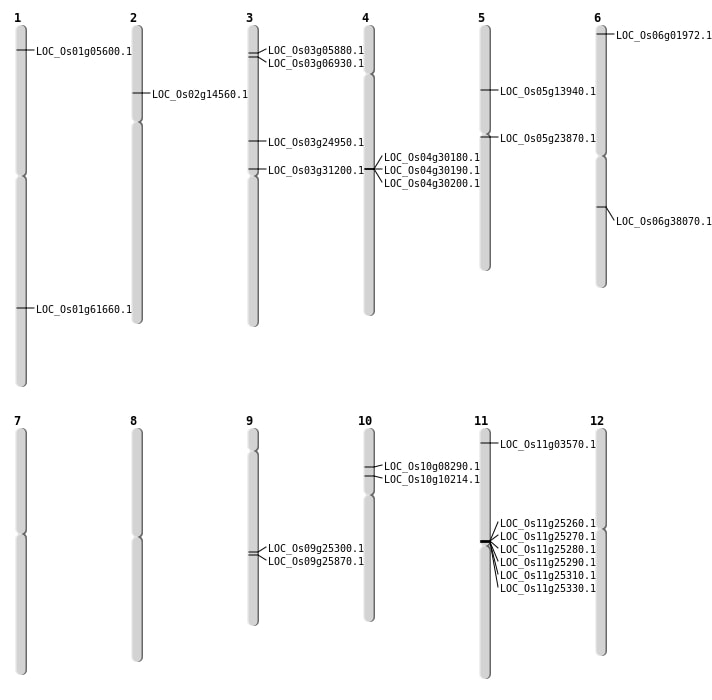
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22137288 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 2670148 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g05600 |
| SBS | Chr1 | 32174517 | C-T | Homo | INTRON | |
| SBS | Chr1 | 40075121 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 5763631 | C-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr1 | 7112249 | T-C | Homo | INTERGENIC | |
| SBS | Chr10 | 366765 | C-T | Homo | UTR_5_PRIME | |
| SBS | Chr10 | 5619500 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g10214 |
| SBS | Chr10 | 8505746 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 14432307 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 14432308 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 20773513 | G-A | Homo | INTERGENIC | |
| SBS | Chr11 | 2487616 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 24242495 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 14235013 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g24950 |
| SBS | Chr3 | 25816885 | A-C | Homo | INTERGENIC | |
| SBS | Chr3 | 30649587 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 26328177 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 7699442 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 7699444 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 7747956 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g13940 |
| SBS | Chr5 | 7773186 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 1087913 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 15066808 | A-G | Homo | INTRON | |
| SBS | Chr6 | 24354419 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 25904990 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 27967439 | G-T | Homo | INTRON | |
| SBS | Chr6 | 547998 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g01972 |
| SBS | Chr7 | 10088171 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 1065828 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 18965796 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 25677377 | T-A | Homo | INTERGENIC | |
| SBS | Chr9 | 12081052 | T-C | Homo | INTRON | |
| SBS | Chr9 | 15514178 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g25870 |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1383156 | 1383158 | 2 | LOC_Os11g03570 |
| Deletion | Chr10 | 1561050 | 1561056 | 6 | |
| Deletion | Chr9 | 1592426 | 1592437 | 11 | |
| Deletion | Chr5 | 2046737 | 2046739 | 2 | |
| Deletion | Chr3 | 3515653 | 3515687 | 34 | LOC_Os03g06930 |
| Deletion | Chr11 | 5674416 | 5674417 | 1 | |
| Deletion | Chr5 | 6613721 | 6613722 | 1 | |
| Deletion | Chr2 | 8026843 | 8026844 | 1 | |
| Deletion | Chr2 | 8027984 | 8027989 | 5 | LOC_Os02g14560 |
| Deletion | Chr10 | 8644405 | 8644418 | 13 | |
| Deletion | Chr7 | 9438733 | 9438742 | 9 | |
| Deletion | Chr12 | 10688067 | 10688086 | 19 | |
| Deletion | Chr3 | 11758987 | 11759001 | 14 | |
| Deletion | Chr6 | 11975337 | 11975348 | 11 | |
| Deletion | Chr5 | 13725937 | 13725948 | 11 | LOC_Os05g23870 |
| Deletion | Chr3 | 13781809 | 13781814 | 5 | |
| Deletion | Chr6 | 13843176 | 13843185 | 9 | |
| Deletion | Chr9 | 14055892 | 14055895 | 3 | |
| Deletion | Chr11 | 14385001 | 14429000 | 44000 | 6 |
| Deletion | Chr9 | 15145204 | 15145293 | 89 | LOC_Os09g25300 |
| Deletion | Chr6 | 15356331 | 15356334 | 3 | |
| Deletion | Chr11 | 15414623 | 15414624 | 1 | |
| Deletion | Chr3 | 17767447 | 17767450 | 3 | LOC_Os03g31200 |
| Deletion | Chr4 | 18014313 | 18025680 | 11367 | 3 |
| Deletion | Chr10 | 21383953 | 21383955 | 2 | |
| Deletion | Chr4 | 22007697 | 22007698 | 1 | |
| Deletion | Chr3 | 22468508 | 22468573 | 65 | |
| Deletion | Chr6 | 22513739 | 22513740 | 1 | LOC_Os06g38070 |
| Deletion | Chr5 | 29074293 | 29074294 | 1 | |
| Deletion | Chr1 | 35674541 | 35674550 | 9 | LOC_Os01g61660 |
| Deletion | Chr1 | 37140312 | 37140314 | 2 | |
| Deletion | Chr1 | 42028750 | 42028755 | 5 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 4481051 | 4481055 | 5 | LOC_Os10g08290 |
| Insertion | Chr12 | 458792 | 458793 | 2 | |
| Insertion | Chr4 | 9530145 | 9530145 | 1 | |
| Insertion | Chr6 | 10109622 | 10109622 | 1 | |
| Insertion | Chr7 | 20205844 | 20205846 | 3 | |
| Insertion | Chr7 | 29146364 | 29146367 | 4 | |
| Insertion | Chr8 | 3234051 | 3234051 | 1 | |
| Insertion | Chr9 | 12945888 | 12945888 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 2718724 | 2951864 | LOC_Os03g05880 |
No Translocation
